Fitness, stress resistance, and extraintestinal virulence in Escherichia coli
- PMID: 23690401
- PMCID: PMC3719556
- DOI: 10.1128/IAI.01329-12
Fitness, stress resistance, and extraintestinal virulence in Escherichia coli
Abstract
The extraintestinal virulence of Escherichia coli is dependent on numerous virulence genes. However, there is growing evidence for a role of the metabolic properties and stress responses of strains in pathogenesis. We assessed the respective roles of these factors in strain virulence by developing phenotypic assays for measuring in vitro individual and competitive fitness and the general stress response, which we applied to 82 commensal and extraintestinal pathogenic E. coli strains previously tested in a mouse model of sepsis. Individual fitness properties, in terms of maximum growth rates in various media (Luria-Bertani broth with and without iron chelator, minimal medium supplemented with gluconate, and human urine) and competitive fitness properties, estimated as the mean relative growth rate per generation in mixed cultures with a reference fluorescent E. coli strain, were highly diverse between strains. The activity of the main general stress response regulator, RpoS, as determined by iodine staining of the colonies, H2O2 resistance, and rpoS sequencing, was also highly variable. No correlation between strain fitness and stress resistance and virulence in the mouse model was found, except that the maximum growth rate in urine was higher for virulent strains. Multivariate analysis showed that the number of virulence factors was the only independent factor explaining the virulence in mice. At the species level, growth capacity and stress resistance are heterogeneous properties that do not contribute significantly to the intrinsic virulence of the strains.
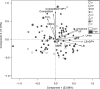
Figures



References
-
- Russo TA, Johnson JR. 2003. Medical and economic impact of extraintestinal infections due to Escherichia coli: focus on an increasingly important endemic problem. Microbes Infect. 5:449–456 - PubMed
-
- Tenaillon O, Skurnik D, Picard B, Denamur E. 2010. The population genetics of commensal Escherichia coli. Nat. Rev. Microbiol. 8:207–217 - PubMed
-
- Desjardins P, Picard B, Kaltenbock B, Elion J, Denamur E. 1995. Sex in Escherichia coli does not disrupt the clonal structure of the population: evidence from random amplified polymorphic DNA and restriction-fragment-length polymorphism. J. Mol. Evol. 41:440–448 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

