A versatile, bar-coded nuclear marker/reporter for live cell fluorescent and multiplexed high content imaging
- PMID: 23691010
- PMCID: PMC3653935
- DOI: 10.1371/journal.pone.0063286
A versatile, bar-coded nuclear marker/reporter for live cell fluorescent and multiplexed high content imaging
Abstract
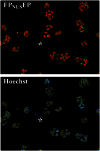
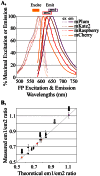
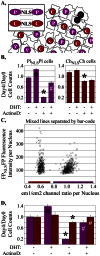
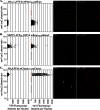
The screening of large numbers of compounds or siRNAs is a mainstay of both academic and pharmaceutical research. Most screens test those interventions against a single biochemical or cellular output whereas recording multiple complementary outputs may be more biologically relevant. High throughput, multi-channel fluorescence microscopy permits multiple outputs to be quantified in specific cellular subcompartments. However, the number of distinct fluorescent outputs available remains limited. Here, we describe a cellular bar-code technology in which multiple cell-based assays are combined in one well after which each assay is distinguished by fluorescence microscopy. The technology uses the unique fluorescent properties of assay-specific markers comprised of distinct combinations of different 'red' fluorescent proteins sandwiched around a nuclear localization signal. The bar-code markers are excited by a common wavelength of light but distinguished ratiometrically by their differing relative fluorescence in two emission channels. Targeting the bar-code to cell nuclei enables individual cells expressing distinguishable markers to be readily separated by standard image analysis programs. We validated the method by showing that the unique responses of different cell-based assays to specific drugs are retained when three assays are co-plated and separated by the bar-code. Based upon those studies, we discuss a roadmap in which even more assays may be combined in a well. The ability to analyze multiple assays simultaneously will enable screens that better identify, characterize and distinguish hits according to multiple biologically or clinically relevant criteria. These capabilities also enable the re-creation of complex mixtures of cell types that is emerging as a central area of interest in many fields.
Conflict of interest statement
Figures




Similar articles
-
Live-cell measurements of kinase activity in single cells using translocation reporters.Nat Protoc. 2018 Jan;13(1):155-169. doi: 10.1038/nprot.2017.128. Epub 2017 Dec 21. Nat Protoc. 2018. PMID: 29266096
-
Quantifying the autophagy-triggering effects of drugs in cell spheroids with live fluorescence microscopy.Methods Mol Biol. 2014;1165:19-29. doi: 10.1007/978-1-4939-0856-1_3. Methods Mol Biol. 2014. PMID: 24839016
-
Highly multiplexed phenotypic imaging for cell proliferation studies.J Biomol Screen. 2014 Jan;19(1):145-57. doi: 10.1177/1087057113495712. Epub 2013 Jul 29. J Biomol Screen. 2014. PMID: 23896684
-
Novel fluorescent proteins for high-content screening.Drug Discov Today. 2006 Dec;11(23-24):1054-60. doi: 10.1016/j.drudis.2006.09.005. Epub 2006 Sep 25. Drug Discov Today. 2006. PMID: 17129823 Review.
-
Photobleaching microscopy reveals the dynamics of mRNA-binding proteins inside live cell nuclei.Prog Mol Subcell Biol. 2004;35:119-34. doi: 10.1007/978-3-540-74266-1_6. Prog Mol Subcell Biol. 2004. PMID: 15113082 Review. No abstract available.
Cited by
-
An optimized genetically encoded dual reporter for simultaneous ratio imaging of Ca2+ and H+ reveals new insights into ion signaling in plants.New Phytol. 2021 Jun;230(6):2292-2310. doi: 10.1111/nph.17202. Epub 2021 Feb 18. New Phytol. 2021. PMID: 33455006 Free PMC article.
-
Maximizing the quantitative accuracy and reproducibility of Förster resonance energy transfer measurement for screening by high throughput widefield microscopy.Methods. 2014 Mar 15;66(2):188-99. doi: 10.1016/j.ymeth.2013.07.040. Epub 2013 Aug 6. Methods. 2014. PMID: 23927839 Free PMC article.
-
A Single-Cell Platform for Monitoring Viral Proteolytic Cleavage in Different Cellular Compartments.Biochem Insights. 2016 Sep 22;8(Suppl 2):23-31. doi: 10.4137/BCI.S30379. eCollection 2015. Biochem Insights. 2016. PMID: 27688710 Free PMC article.
-
Advantages and Limitations of Androgen Receptor-Based Methods for Detecting Anabolic Androgenic Steroid Abuse as Performance Enhancing Drugs.PLoS One. 2016 Mar 21;11(3):e0151860. doi: 10.1371/journal.pone.0151860. eCollection 2016. PLoS One. 2016. PMID: 26998755 Free PMC article.
References
-
- Mayr LM, Bojanic D (2009) Novel trends in high-throughput screening. Curr Opin Pharmacol 9: 580–888. - PubMed
-
- Oheim M (2011) Advances and challenges in high-throughput microscopy for live-cell subcellular imaging. Expert Opin Drug Discov 6: 1299–1315. - PubMed
-
- Krucker T, Sandanaraj BS (2011) Optical imaging for the new grammar of drug discovery. Philos Transact A Math Phys Eng Sci 369: 4651–4665. - PubMed
-
- Carpenter A (2007) Image-based chemical screening. Nat Chem Biol 3: 461–465. - PubMed
-
- Carpenter AE (2009) Extracting rich information from images. Methods Mol Biol 486: 193–211. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

