The minimal Bacillus subtilis nonhomologous end joining repair machinery
- PMID: 23691176
- PMCID: PMC3656841
- DOI: 10.1371/journal.pone.0064232
The minimal Bacillus subtilis nonhomologous end joining repair machinery
Abstract
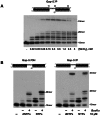
It is widely accepted that repair of double-strand breaks in bacteria that either sporulate or that undergo extended periods of stationary phase relies not only on homologous recombination but also on a minimal nonhomologous end joining (NHEJ) system consisting of a dedicated multifunctional ATP-dependent DNA Ligase D (LigD) and the DNA-end-binding protein Ku. Bacillus subtilis is one of the bacterial members with a NHEJ system that contributes to genome stability during the stationary phase and germination of spores, having been characterized exclusively in vivo. Here, the in vitro analysis of the functional properties of the purified B. subtilis LigD (BsuLigD) and Ku (BsuKu) proteins is presented. The results show that the essential biochemical signatures exhibited by BsuLigD agree with its proposed function in NHEJ: i) inherent polymerization activity showing preferential insertion of NMPs, ii) specific recognition of the phosphate group at the downstream 5' end, iii) intrinsic ligase activity, iv) ability to promote realignments of the template and primer strands during elongation of mispaired 3' ends, and v) it is recruited to DNA by BsuKu that stimulates the inherent polymerization and ligase activities of the enzyme allowing it to deal with and to hold different and unstable DNA realignments.
Conflict of interest statement
Figures



References
-
- Daley JM, Palmbos PL, Wu D, Wilson TE (2005) Nonhomologous end joining in yeast. Annu Rev Genet 39: 431–451. - PubMed
-
- Dudasova Z, Dudas A, Chovanec M (2004) Non-homologous end-joining factors of Saccharomyces cerevisiae . FEMS Microbiol Rev 28: 581–601. - PubMed
-
- Krejci L, Chen L, Van Komen S, Sung P, Tomkinson A (2003) Mending the break: two DNA double-strand break repair machines in eukaryotes. Prog Nucleic Acid Res Mol Biol 74: 159–201. - PubMed
-
- Lees-Miller SP, Meek K (2003) Repair of DNA double strand breaks by non-homologous end joining. Biochimie 85: 1161–1173. - PubMed
-
- Burma S, Chen BP, Chen DJ (2006) Role of non-homologous end joining (NHEJ) in maintaining genomic integrity. DNA Repair (Amst) 5: 1042–1048. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

