Comprehensive dissection of spatiotemporal metabolic shifts in primary, secondary, and lipid metabolism during developmental senescence in Arabidopsis
- PMID: 23696093
- PMCID: PMC3707545
- DOI: 10.1104/pp.113.217380
Comprehensive dissection of spatiotemporal metabolic shifts in primary, secondary, and lipid metabolism during developmental senescence in Arabidopsis
Abstract
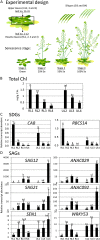
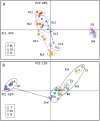
Developmental senescence is a coordinated physiological process in plants and is critical for nutrient redistribution from senescing leaves to newly formed sink organs, including young leaves and developing seeds. Progress has been made concerning the genes involved and the regulatory networks controlling senescence. The resulting complex metabolome changes during senescence have not been investigated in detail yet. Therefore, we conducted a comprehensive profiling of metabolites, including pigments, lipids, sugars, amino acids, organic acids, nutrient ions, and secondary metabolites, and determined approximately 260 metabolites at distinct stages in leaves and siliques during senescence in Arabidopsis (Arabidopsis thaliana). This provided an extensive catalog of metabolites and their spatiotemporal cobehavior with progressing senescence. Comparison with silique data provides clues to source-sink relations. Furthermore, we analyzed the metabolite distribution within single leaves along the basipetal sink-source transition trajectory during senescence. Ceramides, lysolipids, aromatic amino acids, branched chain amino acids, and stress-induced amino acids accumulated, and an imbalance of asparagine/aspartate, glutamate/glutamine, and nutrient ions in the tip region of leaves was detected. Furthermore, the spatiotemporal distribution of tricarboxylic acid cycle intermediates was already changed in the presenescent leaves, and glucosinolates, raffinose, and galactinol accumulated in the base region of leaves with preceding senescence. These results are discussed in the context of current models of the metabolic shifts occurring during developmental and environmentally induced senescence. As senescence processes are correlated to crop yield, the metabolome data and the approach provided here can serve as a blueprint for the analysis of traits and conditions linking crop yield and senescence.
Figures




References
-
- Agati G, Tattini M. (2010) Multiple functional roles of flavonoids in photoprotection. New Phytol 186: 786–793 - PubMed
-
- Alleva R, Tomasetti M, Andera L, Gellert N, Borghi B, Weber C, Murphy MP, Neuzil J. (2001) Coenzyme Q blocks biochemical but not receptor-mediated apoptosis by increasing mitochondrial antioxidant protection. FEBS Lett 503: 46–50 - PubMed
-
- Anderson ME. (1985) Determination of glutathione and glutathione disulfide in biological samples. Methods Enzymol 113: 548–555 - PubMed
-
- Araújo WL, Ishizaki K, Nunes-Nesi A, Larson TR, Tohge T, Krahnert I, Witt S, Obata T, Schauer N, Graham IA, et al (2010) Identification of the 2-hydroxyglutarate and isovaleryl-CoA dehydrogenases as alternative electron donors linking lysine catabolism to the electron transport chain of Arabidopsis mitochondria. Plant Cell 22: 1549–1563 - PMC - PubMed
-
- Araújo WL, Tohge T, Ishizaki K, Leaver CJ, Fernie AR. (2011) Protein degradation: an alternative respiratory substrate for stressed plants. Trends Plant Sci 16: 489–498 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

