Pathogenic mitochondrial tRNA point mutations: nine novel mutations affirm their importance as a cause of mitochondrial disease
- PMID: 23696415
- PMCID: PMC3884772
- DOI: 10.1002/humu.22358
Pathogenic mitochondrial tRNA point mutations: nine novel mutations affirm their importance as a cause of mitochondrial disease
Abstract
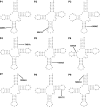
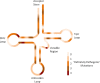
Mutations in the mitochondrial genome, and in particular the mt-tRNAs, are an important cause of human disease. Accurate classification of the pathogenicity of novel variants is vital to allow accurate genetic counseling for patients and their families. The use of weighted criteria based on functional studies-outlined in a validated pathogenicity scoring system--is therefore invaluable in determining whether novel or rare mt-tRNA variants are pathogenic. Here, we describe the identification of nine novel mt--tRNA variants in nine families, in which the probands presented with a diverse range of clinical phenotypes including mitochondrial encephalomyopathy, lactic acidosis, and stroke-like episodes, isolated progressive external ophthalmoplegia, epilepsy, deafness and diabetes. Each of the variants identified (m.4289T>C, MT-TI; m.5541C>T, MT-TW; m.5690A>G, MT-TN; m.7451A>T, MT-TS1; m.7554G>A, MT-TD; m.8304G>A, MT-TK; m.12206C>T, MT-TH; m.12317T>C, MT-TL2; m.16023G>A, MT-TP) was present in a different tRNA, with evidence in support of pathogenicity, and where possible, details of mutation transmission documented. Through the application of the pathogenicity scoring system, we have classified six of these variants as "definitely pathogenic" mutations (m.5541C>T, m.5690A>G, m.7451A>T, m.12206C>T, m.12317T>C, and m.16023G>A), whereas the remaining three currently lack sufficient evidence and are therefore classed as 'possibly pathogenic' (m.4289T>C, m.7554G>A, and m.8304G>A).
Keywords: mitochondrial disease; mitochondrial tRNA; segregation; single-fiber studies.
© 2013 WILEY PERIODICALS, INC.
Figures


References
-
- Crimi M, Galbiati S, Sciacco M, Bordoni A, Natali MG, Raimondi M, Bresolin N, Comi GP. Mitochondrial-DNA nucleotides G4298A and T10010C as pathogenic mutations: the confirmation in two new cases. Mitochondrion. 2004;3(5):279–283. - PubMed
-
- Elson JL, Swalwell H, Blakely EL, McFarland R, Taylor RW, Turnbull DM. Pathogenic mitochondrial tRNA mutations—which mutations are inherited and why? Hum Mutat. 2009;30(11):E984–E992. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

