Diverse neuronal lineages make stereotyped contributions to the Drosophila locomotor control center, the central complex
- PMID: 23696496
- PMCID: PMC3902843
- DOI: 10.1002/cne.23339
Diverse neuronal lineages make stereotyped contributions to the Drosophila locomotor control center, the central complex
Abstract
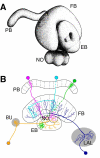
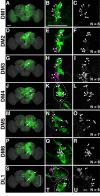
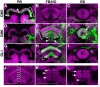
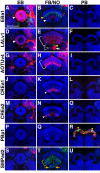
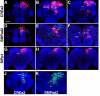
The Drosophila central brain develops from a fixed number of neuroblasts. Each neuroblast makes a clone of neurons that exhibit common trajectories. Here we identified 15 distinct clones that carry larval-born neurons innervating the Drosophila central complex (CX), which consists of four midline structures including the protocerebral bridge (PB), fan-shaped body (FB), ellipsoid body (EB), and noduli (NO). Clonal analysis revealed that the small-field CX neurons, which establish intricate projections across different CX substructures, exist in four isomorphic groups that respectively derive from four complex posterior asense-negative lineages. In terms of the region-characteristic large-field CX neurons, we found that two lineages make PB neurons, 10 lineages produce FB neurons, three lineages generate EB neurons, and two lineages yield NO neurons. The diverse FB developmental origins reflect the discrete input pathways for different FB subcompartments. Clonal analysis enlightens both development and anatomy of the insect locomotor control center.
Copyright © 2013 Wiley Periodicals, Inc.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

