Single-membrane-bounded peroxisome division revealed by isolation of dynamin-based machinery
- PMID: 23696667
- PMCID: PMC3677435
- DOI: 10.1073/pnas.1303483110
Single-membrane-bounded peroxisome division revealed by isolation of dynamin-based machinery
Abstract
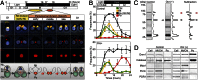
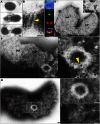
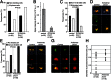
Peroxisomes (microbodies) are ubiquitous single-membrane-bounded organelles and fulfill essential roles in the cellular metabolism. They are found in virtually all eukaryotic cells and basically multiply by division. However, the mechanochemical machinery involved in peroxisome division remains elusive. Here, we first identified the peroxisome-dividing (POD) machinery. We isolated the POD machinery from Cyanidioschyzon merolae, a unicellular red alga containing a single peroxisome. Peroxisomal division in C. merolae can be highly synchronized by light/dark cycles and the microtubule-disrupting agent oryzalin. By proteomic analysis based on the complete genome sequence of C. merolae, we identified a dynamin-related protein 3 (DRP3) ortholog, CmDnm1 (Dnm1), that predominantly accumulated with catalase in the dividing-peroxisome fraction. Immunofluorescence microscopy demonstrated that Dnm1 formed a ring at the division site of the peroxisome. The outlines of the isolated dynamin rings were dimly observed by phase-contrast microscopy and clearly stained for Dnm1. Electron microscopy revealed that the POD machinery was formed at the cytoplasmic side of the equator. Immunoelectron microscopy showed that the POD machinery consisted of an outer dynamin-based ring and an inner filamentous ring. Down-regulation of Dnm1 impaired peroxisomal division. Surprisingly, the same Dnm1 serially controlled peroxisomal division after mitochondrial division. Because genetic deficiencies of Dnm1 orthologs in multiperoxisomal organisms inhibited both mitochondrial and peroxisomal proliferation, it is thought that peroxisomal division by contraction of a dynamin-based machinery is universal among eukaryotes. These findings are useful for understanding the fundamental systems in eukaryotic cells.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Schrader M, Yoon Y. Mitochondria and peroxisomes: Are the ‘big brother’ and the ‘little sister’ closer than assumed? Bioessays. 2007;29(11):1105–1114. - PubMed
-
- Gould SJ, Valle D. Peroxisome biogenesis disorders: Genetics and cell biology. Trends Genet. 2000;16(8):340–345. - PubMed
-
- Koch A, Schneider G, Lüers GH, Schrader M. Peroxisome elongation and constriction but not fission can occur independently of dynamin-like protein 1. J Cell Sci. 2004;117(Pt 17):3995–4006. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

