General mechanism for modulating immunoglobulin effector function
- PMID: 23697368
- PMCID: PMC3683708
- DOI: 10.1073/pnas.1307864110
General mechanism for modulating immunoglobulin effector function
Abstract
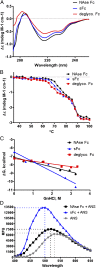
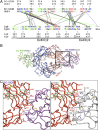
Immunoglobulins recognize and clear microbial pathogens and toxins through the coupling of variable region specificity to Fc-triggered cellular activation. These proinflammatory activities are regulated, thus avoiding the pathogenic sequelae of uncontrolled inflammation by modulating the composition of the Fc-linked glycan. Upon sialylation, the affinities for Fcγ receptors are reduced, whereas those for alternative cellular receptors, such as dendritic cell-specific intercellular adhesion molecule-3-grabbing nonintegrin (DC-SIGN)/CD23, are increased. We demonstrate that sialylation induces significant structural alterations in the Cγ2 domain and propose a model that explains the observed changes in ligand specificity and biological activity. By analogy to related complexes formed by IgE and its evolutionarily related Fc receptors, we conclude that this mechanism is general for the modulation of antibody-triggered immune responses, characterized by a shift between an "open" activating conformation and a "closed" anti-inflammatory state of antibody Fc fragments. This common mechanism has been targeted by pathogens to avoid host defense and offers targets for therapeutic intervention in allergic and autoimmune disorders.
Keywords: conformational change; sialylated IgG Fc.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Comment in
-
Crystal structure of sialylated IgG Fc: implications for the mechanism of intravenous immunoglobulin therapy.Proc Natl Acad Sci U S A. 2013 Sep 17;110(38):E3544-6. doi: 10.1073/pnas.1310657110. Epub 2013 Aug 8. Proc Natl Acad Sci U S A. 2013. PMID: 23929778 Free PMC article. No abstract available.
-
Reply to Crispin et al.: Molecular model that accounts for the biological and physical properties of sialylated Fc.Proc Natl Acad Sci U S A. 2013 Sep 17;110(38):E3547. doi: 10.1073/pnas.1311721110. Proc Natl Acad Sci U S A. 2013. PMID: 24171201 Free PMC article. No abstract available.
References
-
- Sondermann P, Huber R, Oosthuizen V, Jacob U. The 3.2-A crystal structure of the human IgG1 Fc fragment-Fc gammaRIII complex. Nature. 2000;406(6793):267–273. - PubMed
-
- Garman SC, Wurzburg BA, Tarchevskaya SS, Kinet JP, Jardetzky TS. Structure of the Fc fragment of human IgE bound to its high-affinity receptor Fc epsilonRI alpha. Nature. 2000;406(6793):259–266. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

