The roles of 3'-exoribonucleases and the exosome in trypanosome mRNA degradation
- PMID: 23697549
- PMCID: PMC3683928
- DOI: 10.1261/rna.038430.113
The roles of 3'-exoribonucleases and the exosome in trypanosome mRNA degradation
Abstract
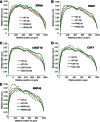
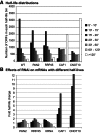
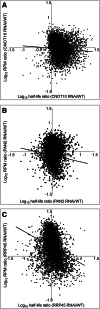
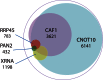
The degradation of eukaryotic mRNAs can be initiated by deadenylation, decapping, or endonuclease cleavage. This is followed by 5'-3' degradation by homologs of Xrn1, and/or 3'-5' degradation by the exosome. We previously reported that, in African trypanosome Trypanosoma brucei, most mRNAs are deadenylated prior to degradation, and that depletion of the major 5'-3' exoribonuclease XRNA preferentially stabilizes unstable mRNAs. We now show that depletion of either CAF1 or CNOT10, two components of the principal deadenylation complex, strongly inhibits degradation of most mRNAs. RNAi targeting another deadenylase, PAN2, or RRP45, a core component of the exosome, preferentially stabilized mRNAs with intermediate half-lives. RRP45 depletion resulted in a 5' bias of mRNA sequences, suggesting action by a distributive 3'-5' exoribonuclease. Results suggested that the exosome is involved in the processing of trypanosome snoRNAs. There was no correlation between effects on half-lives and on mRNA abundance.
Keywords: CAF1; PAN2; Trypanosoma; deadenylation; exosome; mRNA decay; mRNA degradation.
Figures



References
-
- Alibu VP, Storm L, Haile S, Clayton C, Horn D 2004. A doubly inducible system for RNA interference and rapid RNAi plasmid construction in Trypanosoma brucei. Mol Biochem Parasitol 139: 75–82 - PubMed
-
- Banerjee S, Pedersen T 2003. The design, implementation and use of the Ngram statistic package. In Proceedings of the fourth international conference on intelligent text processing and comparative linguistics, 370–381, Mexico City
-
- Banerjee H, Palenchar J, Lukaszewicz M, Bojarska E, Stepinski J, Jemielity J, Guranowski A, Ng S, Wah D, Darzynkiewicz E, et al. 2009. Identification of the HIT-45 protein from Trypanosoma brucei as an FHIT protein/dinucleoside triphosphatase: Substrate specificity studies on the recombinant and endogenous proteins. RNA 15: 1554–1564 - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
