Footprint of APOBEC3 on the genome of human retroelements
- PMID: 23698293
- PMCID: PMC3700199
- DOI: 10.1128/JVI.00298-13
Footprint of APOBEC3 on the genome of human retroelements
Abstract
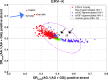
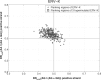
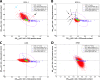
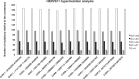
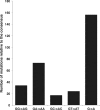
Almost half of the human genome is composed of transposable elements. The genomic structures and life cycles of some of these elements suggest they are a result of waves of retroviral infection and transposition over millions of years. The reduction of retrotransposition activity in primates compared to that in nonprimates, such as mice, has been attributed to the positive selection of several antiretroviral factors, such as apolipoprotein B mRNA editing enzymes. Among these, APOBEC3G is known to mutate G to A within the context of GG in the genome of endogenous as well as several exogenous retroelements (the underlining marks the G that is mutated). On the other hand, APOBEC3F and to a lesser extent other APOBEC3 members induce G-to-A changes within the nucleotide GA. It is known that these enzymes can induce deleterious mutations in the genome of retroviral sequences, but the evolution and/or inactivation of retroelements as a result of mutation by these proteins is not clear. Here, we analyze the mutation signatures of these proteins on large populations of long interspersed nuclear element (LINE), short interspersed nuclear element (SINE), and endogenous retrovirus (ERV) families in the human genome to infer possible evolutionary pressure and/or hypermutation events. Sequence context dependency of mutation by APOBEC3 allows investigation of the changes in the genome of retroelements by inspecting the depletion of G and enrichment of A within the APOBEC3 target and product motifs, respectively. Analysis of approximately 22,000 LINE-1 (L1), 24,000 SINE Alu, and 3,000 ERV sequences showed a footprint of GG→AG mutation by APOBEC3G and GA→AA mutation by other members of the APOBEC3 family (e.g., APOBEC3F) on the genome of ERV-K and ERV-1 elements but not on those of ERV-L, LINE, or SINE.
Figures




References
-
- Kazazian HH., Jr 2004. Mobile elements: drivers of genome evolution. Science 303:1626–1632 - PubMed
-
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409:860–921 - PubMed
-
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, Antonarakis SE, Attwood J, Baertsch R, Bailey J, Barlow K, Beck S, Berry E, Birren B, Bloom T, Bork P, Botcherby M, Bray N, Brent MR, Brown DG, Brown SD, Bult C, Burton J, Butler J, Campbell RD, Carninci P, Cawley S, Chiaromonte F, Chinwalla AT, Church DM, Clamp M, Clee C, Collins FS, Cook LL, Copley RR, Coulson A, Couronne O, Cuff J, Curwen V, Cutts T, Daly M, David R, Davies J, Delehaunty KD, Deri J, Dermitzakis ET, et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420:520–562 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

