Different requirements for scavenger receptor class B type I in hepatitis C virus cell-free versus cell-to-cell transmission
- PMID: 23698298
- PMCID: PMC3719822
- DOI: 10.1128/JVI.01102-13
Different requirements for scavenger receptor class B type I in hepatitis C virus cell-free versus cell-to-cell transmission
Abstract
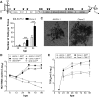
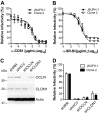
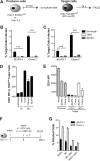
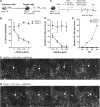
Hepatitis C virus (HCV) is believed to initially infect the liver through the basolateral side of hepatocytes, where it engages attachment factors and the coreceptors CD81 and scavenger receptor class B type I (SR-BI). Active transport toward the apical side brings the virus in close proximity of additional entry factors, the tight junction molecules claudin-1 and occludin. HCV is also thought to propagate via cell-to-cell spread, which allows highly efficient virion delivery to neighboring cells. In this study, we compared an adapted HCV genome, clone 2, characterized by superior cell-to cell spread, to its parental genome, J6/JFH-1, with the goal of elucidating the molecular mechanisms of HCV cell-to-cell transmission. We show that CD81 levels on the donor cells influence the efficiency of cell-to-cell spread and CD81 transfer between neighboring cells correlates with the capacity of target cells to become infected. Spread of J6/JFH-1 was blocked by anti-SR-BI antibody or in cells knocked down for SR-BI, suggesting a direct role for this receptor in HCV cell-to-cell transmission. In contrast, clone 2 displayed a significantly reduced dependence on SR-BI for lateral spread. Mutations in E1 and E2 responsible for the enhanced cell-to-cell spread phenotype of clone 2 rendered cell-free virus more susceptible to antibody-mediated neutralization. Our results indicate that although HCV can lose SR-BI dependence for cell-to-cell spread, vulnerability to neutralizing antibodies may limit this evolutionary option in vivo. Combination therapies targeting both the HCV glycoproteins and SR-BI may therefore hold promise for effective control of HCV dissemination.
Figures

References
-
- Lavanchy D. 2011. Evolving epidemiology of hepatitis C virus. Clin. Microbiol. Infect. 17:107–115 - PubMed
-
- Zeisel MB, Cosset FL, Baumert TF. 2008. Host neutralizing responses and pathogenesis of hepatitis C virus infection. Hepatology 48:299–307 - PubMed
-
- Maillard P, Huby T, Andreo U, Moreau M, Chapman J, Budkowska A. 2006. The interaction of natural hepatitis C virus with human scavenger receptor SR-BI/ClaI is mediated by ApoB-containing lipoproteins. FASEB J. 20:735–737 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

