Role of the phosphatidylserine receptor TIM-1 in enveloped-virus entry
- PMID: 23698310
- PMCID: PMC3719829
- DOI: 10.1128/JVI.01025-13
Role of the phosphatidylserine receptor TIM-1 in enveloped-virus entry
Abstract
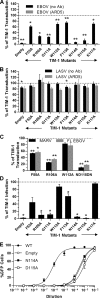
The cell surface receptor T cell immunoglobulin mucin domain 1 (TIM-1) dramatically enhances filovirus infection of epithelial cells. Here, we showed that key phosphatidylserine (PtdSer) binding residues of the TIM-1 IgV domain are critical for Ebola virus (EBOV) entry through direct interaction with PtdSer on the viral envelope. PtdSer liposomes but not phosphatidylcholine liposomes competed with TIM-1 for EBOV pseudovirion binding and transduction. Further, annexin V (AnxV) substituted for the TIM-1 IgV domain, supporting a PtdSer-dependent mechanism. Our findings suggest that TIM-1-dependent uptake of EBOV occurs by apoptotic mimicry. Additionally, TIM-1 enhanced infection of a wide range of enveloped viruses, including alphaviruses and a baculovirus. As further evidence of the critical role of enveloped-virion-associated PtdSer in TIM-1-mediated uptake, TIM-1 enhanced internalization of pseudovirions and virus-like proteins (VLPs) lacking a glycoprotein, providing evidence that TIM-1 and PtdSer-binding receptors can mediate virus uptake independent of a glycoprotein. These results provide evidence for a broad role of TIM-1 as a PtdSer-binding receptor that mediates enveloped-virus uptake. Utilization of PtdSer-binding receptors may explain the wide tropism of many of these viruses and provide new avenues for controlling their virulence.
Figures




References
-
- Leroy EM, Gonzalez JP, Baize S. 2011. Ebola and Marburg haemorrhagic fever viruses: major scientific advances, but a relatively minor public health threat for Africa. Clin. Microbiol. Infect. 17:964–976 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

