Organization of the ER-Golgi interface for membrane traffic control
- PMID: 23698585
- PMCID: PMC4064004
- DOI: 10.1038/nrm3588
Organization of the ER-Golgi interface for membrane traffic control
Abstract
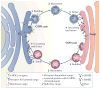
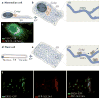
Coat protein complex I (COPI) and COPII are required for bidirectional membrane trafficking between the endoplasmic reticulum (ER) and the Golgi. While these core coat machineries and other transport factors are highly conserved across species, high-resolution imaging studies indicate that the organization of the ER-Golgi interface is varied in eukaryotic cells. Regulation of COPII assembly, in some cases to manage distinct cellular cargo, is emerging as one important component in determining this structure. Comparison of the ER-Golgi interface across different systems, particularly mammalian and plant cells, reveals fundamental elements and distinct organization of this interface. A better understanding of how these interfaces are regulated to meet varying cellular secretory demands should provide key insights into the mechanisms that control efficient trafficking of proteins and lipids through the secretory pathway.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

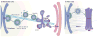
Comment in
-
ER-Golgi transport: authors' response.Nat Rev Mol Cell Biol. 2014 Mar;15(3):1. doi: 10.1038/nrm3588-c2. Epub 2014 Feb 5. Nat Rev Mol Cell Biol. 2014. PMID: 24496388 No abstract available.
-
ER-Golgi transport could occur in the absence of COPII vesicles.Nat Rev Mol Cell Biol. 2014 Mar;15(3):1. doi: 10.1038/nrm3588-c1. Epub 2014 Feb 5. Nat Rev Mol Cell Biol. 2014. PMID: 24496389 No abstract available.
References
-
- Altan-Bonnet N, Sougrat R, Lippincott-Schwartz J. Molecular basis for Golgi maintenance and biogenesis. Curr Opin Cell Biol. 2004;16:364–372. - PubMed
-
- Driouich A, Staehelin LA. In: The Golgi Apparatus. Berger EG, Roth J, editors. Birkhäuser Verlag; 1997. pp. 275–301.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

