Assembling the 20 Gb white spruce (Picea glauca) genome from whole-genome shotgun sequencing data
- PMID: 23698863
- PMCID: PMC3673215
- DOI: 10.1093/bioinformatics/btt178
Assembling the 20 Gb white spruce (Picea glauca) genome from whole-genome shotgun sequencing data
Abstract
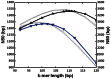
White spruce (Picea glauca) is a dominant conifer of the boreal forests of North America, and providing genomics resources for this commercially valuable tree will help improve forest management and conservation efforts. Sequencing and assembling the large and highly repetitive spruce genome though pushes the boundaries of the current technology. Here, we describe a whole-genome shotgun sequencing strategy using two Illumina sequencing platforms and an assembly approach using the ABySS software. We report a 20.8 giga base pairs draft genome in 4.9 million scaffolds, with a scaffold N50 of 20,356 bp. We demonstrate how recent improvements in the sequencing technology, especially increasing read lengths and paired end reads from longer fragments have a major impact on the assembly contiguity. We also note that scalable bioinformatics tools are instrumental in providing rapid draft assemblies.
Availability: The Picea glauca genome sequencing and assembly data are available through NCBI (Accession#: ALWZ0100000000 PID: PRJNA83435). http://www.ncbi.nlm.nih.gov/bioproject/83435.
Figures
Comment in
-
Genomics: Sprucing up forest tree genomics.Nat Rev Genet. 2013 Jul;14(7):444. doi: 10.1038/nrg3518. Epub 2013 Jun 4. Nat Rev Genet. 2013. PMID: 23732336 No abstract available.
References
-
- Altschul SF, et al. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. - PubMed
-
- Burrows M, Wheeler D. Technical Report 124. Palo Alto, CA: Digital Equipment Corporation; 1994. A block sorting lossless data compression algorithm.
Publication types
MeSH terms
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

