Cells surviving fractional killing by TRAIL exhibit transient but sustainable resistance and inflammatory phenotypes
- PMID: 23699397
- PMCID: PMC3708725
- DOI: 10.1091/mbc.E12-10-0737
Cells surviving fractional killing by TRAIL exhibit transient but sustainable resistance and inflammatory phenotypes
Abstract
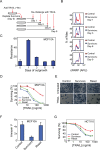
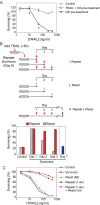
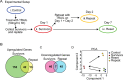
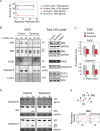
When clonal populations of human cells are exposed to apoptosis-inducing agents, some cells die and others survive. This fractional killing arises not from mutation but from preexisting, stochastic differences in the levels and activities of proteins regulating apoptosis. Here we examine the properties of cells that survive treatment with agonists of two distinct death receptors, tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) and anti-FasR antibodies. We find that "survivor" cells are highly resistant to a second ligand dose applied 1 d later. Resistance is reversible, resetting after several days of culture in the absence of death ligand. "Reset" cells appear identical to drug-naive cells with respect to death ligand sensitivity and gene expression profiles. TRAIL survivors are cross-resistant to activators of FasR and vice versa and exhibit an NF-κB-dependent inflammatory phenotype. Remarkably, reversible resistance is induced in the absence of cell death when caspase inhibitors are present and can be sustained for 1 wk or more, also without cell death, by periodic ligand exposure. Thus stochastic differences in cell state can have sustained consequences for sen-sitivity to prodeath ligands and acquisition of proinflammatory phenotypes. The important role played by periodicity in TRAIL exposure for induction of opposing apoptosis and survival mechanisms has implications for the design of optimal therapeutic agents and protocols.
Figures



References
-
- Adams C, et al. Structural and functional analysis of the interaction between the agonistic monoclonal antibody Apomab and the proapoptotic receptor DR5. Cell Death Differ. 2008;15:751–761. - PubMed
-
- Ashkenazi A. Targeting the extrinsic apoptosis pathway in cancer. Cytokine Growth Factor Rev. 2008;19:325–331. - PubMed
-
- Ashkenazi A, Dixit VM. Apoptosis control by death and decoy receptors. Curr Opin Cell Biol. 1999;11:255–260. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

