Mule/Huwe1/Arf-BP1 suppresses Ras-driven tumorigenesis by preventing c-Myc/Miz1-mediated down-regulation of p21 and p15
- PMID: 23699408
- PMCID: PMC3672645
- DOI: 10.1101/gad.214577.113
Mule/Huwe1/Arf-BP1 suppresses Ras-driven tumorigenesis by preventing c-Myc/Miz1-mediated down-regulation of p21 and p15
Abstract
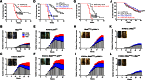
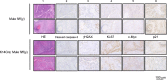
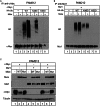
Tumorigenesis results from dysregulation of oncogenes and tumor suppressors that influence cellular proliferation, differentiation, apoptosis, and/or senescence. Many gene products involved in these processes are substrates of the E3 ubiquitin ligase Mule/Huwe1/Arf-BP1 (Mule), but whether Mule acts as an oncogene or tumor suppressor in vivo remains controversial. We generated K14Cre;Mule(flox/flox(y)) (Mule kKO) mice and subjected them to DMBA/PMA-induced skin carcinogenesis, which depends on oncogenic Ras signaling. Mule deficiency resulted in increased penetrance, number, and severity of skin tumors, which could be reversed by concomitant genetic knockout of c-Myc but not by knockout of p53 or p19Arf. Notably, in the absence of Mule, c-Myc/Miz1 transcriptional complexes accumulated, and levels of p21CDKN1A (p21) and p15INK4B (p15) were down-regulated. In vitro, Mule-deficient primary keratinocytes exhibited increased proliferation that could be reversed by Miz1 knockdown. Transfer of Mule-deficient transformed cells to nude mice resulted in enhanced tumor growth that again could be abrogated by Miz1 knockdown. Our data demonstrate in vivo that Mule suppresses Ras-mediated tumorigenesis by preventing an accumulation of c-Myc/Miz1 complexes that mediates p21 and p15 down-regulation.
Keywords: Huwe1; Miz1; Mule; Ras; c-Myc; p21.
Figures




Similar articles
-
Miz1 is a critical repressor of cdkn1a during skin tumorigenesis.PLoS One. 2012;7(4):e34885. doi: 10.1371/journal.pone.0034885. Epub 2012 Apr 11. PLoS One. 2012. PMID: 22509363 Free PMC article.
-
The MULE/HUWE1 E3 ubiquitin ligase is a tumor suppressor.Cancer Discov. 2013 Jul;3(7):OF32. doi: 10.1158/2159-8290.CD-RW2013-119. Epub 2013 Jun 6. Cancer Discov. 2013. PMID: 23847377
-
Miz1 and HectH9 regulate the stability of the checkpoint protein, TopBP1.EMBO J. 2008 Nov 5;27(21):2851-61. doi: 10.1038/emboj.2008.200. Epub 2008 Oct 16. EMBO J. 2008. PMID: 18923429 Free PMC article.
-
c-Myc and activated Ras during skin tumorigenesis: cooperation at the cancer stem cell level?Ernst Schering Found Symp Proc. 2006;(5):13-26. doi: 10.1007/2789_2007_042. Ernst Schering Found Symp Proc. 2006. PMID: 17939293 Review.
-
Alterations in epidermal biochemistry as a consequence of stage-specific genetic changes in skin carcinogenesis.Environ Health Perspect. 1991 Jun;93:3-10. doi: 10.1289/ehp.91933. Environ Health Perspect. 1991. PMID: 1773799 Free PMC article. Review.
Cited by
-
The ubiquitin ligase Huwe1 regulates the maintenance and lymphoid commitment of hematopoietic stem cells.Nat Immunol. 2016 Nov;17(11):1312-1321. doi: 10.1038/ni.3559. Epub 2016 Sep 26. Nat Immunol. 2016. PMID: 27668798 Free PMC article.
-
Writing and erasing MYC ubiquitination and SUMOylation.Genes Dis. 2019 Jul 24;6(4):359-371. doi: 10.1016/j.gendis.2019.05.006. eCollection 2019 Dec. Genes Dis. 2019. PMID: 31832515 Free PMC article. Review.
-
Mcl-1 ubiquitination: unique regulation of an essential survival protein.Cells. 2014 May 8;3(2):418-37. doi: 10.3390/cells3020418. Cells. 2014. PMID: 24814761 Free PMC article.
-
The IDH2 R172K mutation associated with angioimmunoblastic T-cell lymphoma produces 2HG in T cells and impacts lymphoid development.Proc Natl Acad Sci U S A. 2016 Dec 27;113(52):15084-15089. doi: 10.1073/pnas.1617929114. Epub 2016 Dec 12. Proc Natl Acad Sci U S A. 2016. PMID: 27956631 Free PMC article.
-
MEK1 is required for the development of NRAS-driven leukemia.Oncotarget. 2016 Dec 6;7(49):80113-80130. doi: 10.18632/oncotarget.12555. Oncotarget. 2016. PMID: 27741509 Free PMC article.
References
-
- Adhikary S, Marinoni F, Hock A, Hulleman E, Popov N, Beier R, Bernard S, Quarto M, Capra M, Goettig S, et al. 2005. The ubiquitin ligase HectH9 regulates transcriptional activation by Myc and is essential for tumor cell proliferation. Cell 123: 409–421 - PubMed
-
- Bos JL 1989. ras oncogenes in human cancer: A review. Cancer Res 49: 4682–4689 - PubMed
-
- Bringold F, Serrano M 2000. Tumor suppressors and oncogenes in cellular senescence. Exp Gerontol 35: 317–329 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
