Functional implication of β-carotene hydroxylases in soybean nodulation
- PMID: 23700351
- PMCID: PMC3707551
- DOI: 10.1104/pp.113.215020
Functional implication of β-carotene hydroxylases in soybean nodulation
Abstract
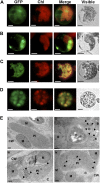
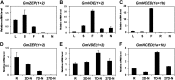
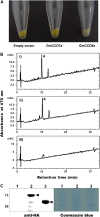
Legume-Rhizobium spp. symbiosis requires signaling between the symbiotic partners and differential expression of plant genes during nodule development. Previously, we cloned a gene encoding a putative β-carotene hydroxylase (GmBCH1) from soybean (Glycine max) whose expression increased during nodulation with Bradyrhizobium japonicum. In this work, we extended our study to three GmBCHs to examine their possible role(s) in nodule development, as they were additionally identified as nodule specific, along with the completion of the soybean genome. In situ hybridization revealed the expression of three GmBCHs (GmBCH1, GmBCH2, and GmBCH3) in the infected cells of root nodules, and their enzymatic activities were confirmed by functional assays in Escherichia coli. Localization of GmBCHs by transfecting Arabidopsis (Arabidopsis thaliana) protoplasts with green fluorescent protein fusions and by electron microscopic immunogold detection in soybean nodules indicated that GmBCH2 and GmBCH3 were present in plastids, while GmBCH1 appeared to be cytosolic. RNA interference of the GmBCHs severely impaired nitrogen fixation as well as nodule development. Surprisingly, we failed to detect zeaxanthin, a product of GmBCH, or any other carotenoids in nodules. Therefore, we examined the possibility that most of the carotenoids in nodules are converted or cleaved to other compounds. We detected the expression of some carotenoid cleavage dioxygenases (GmCCDs) in wild-type nodules and also a reduced amount of zeaxanthin in GmCCD8-expressing E. coli, suggesting cleavage of the carotenoid. In view of these findings, we propose that carotenoids such as zeaxanthin synthesized in root nodules are cleaved by GmCCDs, and we discuss the possible roles of the carotenoid cleavage products in nodulation.
Figures





References
-
- Alder A, Holdermann I, Beyer P, Al-Babili S. (2008) Carotenoid oxygenases involved in plant branching catalyse a highly specific conserved apocarotenoid cleavage reaction. Biochem J 416: 289–296 - PubMed
-
- Auldridge ME, Block A, Vogel JT, Dabney-Smith C, Mila I, Bouzayen M, Magallanes-Lundback M, DellaPenna D, McCarty DR, Klee HJ. (2006a) Characterization of three members of the Arabidopsis carotenoid cleavage dioxygenase family demonstrates the divergent roles of this multifunctional enzyme family. Plant J 45: 982–993 - PubMed
-
- Auldridge ME, McCarty DR, Klee HJ. (2006b) Plant carotenoid cleavage oxygenases and their apocarotenoid products. Curr Opin Plant Biol 9: 315–321 - PubMed
-
- Booker J, Auldridge M, Wills S, McCarty D, Klee H, Leyser O. (2004) MAX3/CCD7 is a carotenoid cleavage dioxygenase required for the synthesis of a novel plant signaling molecule. Curr Biol 14: 1232–1238 - PubMed
-
- Bouvier F, Isner JC, Dogbo O, Camara B. (2005) Oxidative tailoring of carotenoids: a prospect towards novel functions in plants. Trends Plant Sci 10: 187–194 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

