Insights into organ-specific pathogen defense responses in plants: RNA-seq analysis of potato tuber-Phytophthora infestans interactions
- PMID: 23702331
- PMCID: PMC3674932
- DOI: 10.1186/1471-2164-14-340
Insights into organ-specific pathogen defense responses in plants: RNA-seq analysis of potato tuber-Phytophthora infestans interactions
Abstract
Background: The late blight pathogen Phytophthora infestans can attack both potato foliage and tubers. Although interaction transcriptome dynamics between potato foliage and various pathogens have been reported, no transcriptome study has focused specifically upon how potato tubers respond to pathogen infection. When inoculated with P. infestans, tubers of nontransformed 'Russet Burbank' (WT) potato develop late blight disease while those of transgenic 'Russet Burbank' line SP2211 (+RB), which expresses the potato late blight resistance gene RB (Rpi-blb1), do not. We compared transcriptome responses to P. infestans inoculation in tubers of these two lines.
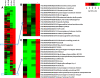
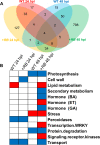
Results: We demonstrated the practicality of RNA-seq to study tetraploid potato and present the first RNA-seq study of potato tuber diseases. A total of 483 million paired end Illumina RNA-seq reads were generated, representing the transcription of around 30,000 potato genes. Differentially expressed genes, gene groups and ontology bins that exhibited differences between the WT and +RB lines were identified. P. infestans transcripts, including those of known effectors, were also identified.
Conclusion: Faster and stronger activation of defense related genes, gene groups and ontology bins correlate with successful tuber resistance against P. infestans. Our results suggest that the hypersensitive response is likely a general form of resistance against the hemibiotrophic P. infestans-even in potato tubers, organs that develop below ground.
Figures




References
-
- Stevenson WR, Loria R, Frank GD, Weingartner DP. Compedium of potato diseases. Saint Paul, MN, USA: APS Press; 2001.
-
- Kirk WW, Felcher KJ, Douches DS, Niemira BA, Hammerschmidt R. Susceptibility of potato (Solanum tuberosum L.) foliage and tubers to the US8 genotype of Phytophthora infestans. Am J Potato Res. 2001;78(4):319–322. doi: 10.1007/BF02875697. - DOI
-
- Song JQ, Bradeen JM, Naess SK, Raasch JA, Wielgus SM, Haberlach GT, Liu J, Kuang HH, Austin-Phillips S, Buell CR. Gene RB cloned from Solanum bulbocastanum confers broad spectrum resistance to potato late blight. Proc Natl Acad Sci USA. 2003;100(16):9128–9133. doi: 10.1073/pnas.1533501100. - DOI - PMC - PubMed
-
- van der Vossen E, Sikkema A, Hekkert BTL, Gros J, Stevens P, Muskens M, Wouters D, Pereira A, Stiekema W, Allefs S. An ancient R gene from the wild potato species Solanum bulbocastanum confers broad-spectrum resistance to Phytophthora infestans in cultivated potato and tomato. Plant J. 2003;36(6):867–882. doi: 10.1046/j.1365-313X.2003.01934.x. - DOI - PubMed
-
- Bradeen JM, Lorizzo M, Mollov DS, Raasch J, Kramer LC, Millett BP, Austin-Phillips S, Jiang JM, Carputo D. Higher copy numbers of the potato RB transgene correspond to enhanced transcript and late blight resistance levels. Mol Plant Microbe Interact. 2009;22(4):437–446. doi: 10.1094/MPMI-22-4-0437. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

