Community transcriptomic assembly reveals microbes that contribute to deep-sea carbon and nitrogen cycling
- PMID: 23702516
- PMCID: PMC3965313
- DOI: 10.1038/ismej.2013.85
Community transcriptomic assembly reveals microbes that contribute to deep-sea carbon and nitrogen cycling
Erratum in
- ISME J. 2013 Oct;7(10):2060
Abstract
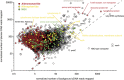
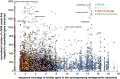
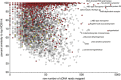
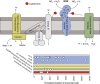
The deep ocean is an important component of global biogeochemical cycles because it contains one of the largest pools of reactive carbon and nitrogen on earth. However, the microbial communities that drive deep-sea geochemistry are vastly unexplored. Metatranscriptomics offers new windows into these communities, but it has been hampered by reliance on genome databases for interpretation. We reconstructed the transcriptomes of microbial populations from Guaymas Basin, in the deep Gulf of California, through shotgun sequencing and de novo assembly of total community RNA. Many of the resulting messenger RNA (mRNA) contiguous sequences contain multiple genes, reflecting co-transcription of operons, including those from dominant members. Also prevalent were transcripts with only limited representation (2.8 times coverage) in a corresponding metagenome, including a considerable portion (1.2 Mb total assembled mRNA sequence) with similarity (96%) to a marine heterotroph, Alteromonas macleodii. This Alteromonas and euryarchaeal marine group II populations displayed abundant transcripts from amino-acid transporters, suggesting recycling of organic carbon and nitrogen from amino acids. Also among the most abundant mRNAs were catalytic subunits of the nitrite oxidoreductase complex and electron transfer components involved in nitrite oxidation. These and other novel genes are related to novel Nitrospirae and have limited representation in accompanying metagenomic data. High throughput sequencing of 16S ribosomal RNA (rRNA) genes and rRNA read counts confirmed that Nitrospirae are minor yet widespread members of deep-sea communities. These results implicate a novel bacterial group in deep-sea nitrite oxidation, the second step of nitrification. This study highlights metatranscriptomic assembly as a valuable approach to study microbial communities.
Figures


References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Bock E, Koops HP, Harms H, Ahlers B.1991. In: Shively JM, Barton LL, (eds)Variations in Autotrophic Life Academic Press: London; 171–200.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

