Analysis of disease-causing GATA1 mutations in murine gene complementation systems
- PMID: 23704091
- PMCID: PMC3695365
- DOI: 10.1182/blood-2013-03-488080
Analysis of disease-causing GATA1 mutations in murine gene complementation systems
Abstract
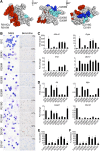
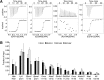
Missense mutations in transcription factor GATA1 underlie a spectrum of congenital red blood cell and platelet disorders. We investigated how these alterations cause distinct clinical phenotypes by combining structural, biochemical, and genomic approaches with gene complementation systems that examine GATA1 function in biologically relevant cellular contexts. Substitutions that disrupt FOG1 cofactor binding impair both gene activation and repression and are associated with pronounced clinical phenotypes. Moreover, clinical severity correlates with the degree of FOG1 disruption. Surprisingly, 2 mutations shown to impair DNA binding of GATA1 in vitro did not measurably affect in vivo target gene occupancy. Rather, one of these disrupted binding to the TAL1 complex, implicating it in diseases caused by GATA1 mutations. Diminished TAL1 complex recruitment mainly impairs transcriptional activation and is linked to relatively mild disease. Notably, different substitutions at the same amino acid can selectively inhibit TAL1 complex or FOG1 binding, producing distinct cellular and clinical phenotypes. The structure-function relationships elucidated here were not predicted by prior in vitro or computational studies. Thus, our findings uncover novel disease mechanisms underlying GATA1 mutations and highlight the power of gene complementation assays for elucidating the molecular basis of genetic diseases.
Figures



References
-
- Martin DI, Orkin SH. Transcriptional activation and DNA binding by the erythroid factor GF-1/NF-E1/Eryf 1. Genes Dev. 1990;4(11):1886–1898. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

