Transcription factors MYOCD, SRF, Mesp1 and SMARCD3 enhance the cardio-inducing effect of GATA4, TBX5, and MEF2C during direct cellular reprogramming
- PMID: 23704920
- PMCID: PMC3660533
- DOI: 10.1371/journal.pone.0063577
Transcription factors MYOCD, SRF, Mesp1 and SMARCD3 enhance the cardio-inducing effect of GATA4, TBX5, and MEF2C during direct cellular reprogramming
Abstract
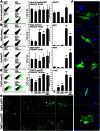
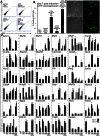
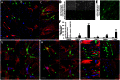
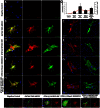
Transient overexpression of defined combinations of master regulator genes can effectively induce cellular reprogramming: the acquisition of an alternative predicted phenotype from a differentiated cell lineage. This can be of particular importance in cardiac regenerative medicine wherein the heart lacks the capacity to heal itself, but simultaneously contains a large pool of fibroblasts. In this study we determined the cardio-inducing capacity of ten transcription factors to actuate cellular reprogramming of mouse embryonic fibroblasts into cardiomyocyte-like cells. Overexpression of transcription factors MYOCD and SRF alone or in conjunction with Mesp1 and SMARCD3 enhanced the basal but necessary cardio-inducing effect of the previously reported GATA4, TBX5, and MEF2C. In particular, combinations of five or seven transcription factors enhanced the activation of cardiac reporter vectors, and induced an upregulation of cardiac-specific genes. Global gene expression analysis also demonstrated a significantly greater cardio-inducing effect when the transcription factors MYOCD and SRF were used. Detection of cross-striated cells was highly dependent on the cell culture conditions and was enhanced by the addition of valproic acid and JAK inhibitor. Although we detected Ca(2+) transient oscillations in the reprogrammed cells, we did not detect significant changes in resting membrane potential or spontaneously contracting cells. This study further elucidates the cardio-inducing effect of the transcriptional networks involved in cardiac cellular reprogramming, contributing to the ongoing rational design of a robust protocol required for cardiac regenerative therapies.
Conflict of interest statement
Figures

References
-
- Murry CE, Reinecke H, Pabon LM (2006) Regeneration gaps: observations on stem cells and cardiac repair. Journal of the American College of Cardiology 47: 1777–1785. - PubMed
-
- Maltsev VA, Rohwedel J, Hescheler J, Wobus AM (1993) Embryonic stem cells differentiate in vitro into cardiomyocytes representing sinusnodal, atrial and ventricular cell types. Mechanisms of development 44: 41–50. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

