Evolution of smooth tubercle Bacilli PE and PE_PGRS genes: evidence for a prominent role of recombination and imprint of positive selection
- PMID: 23705005
- PMCID: PMC3660525
- DOI: 10.1371/journal.pone.0064718
Evolution of smooth tubercle Bacilli PE and PE_PGRS genes: evidence for a prominent role of recombination and imprint of positive selection
Abstract
Background: PE and PE_PGRS are two mycobateria-restricted multigene families encoding membrane associated and secreted proteins that have expanded mainly in the pathogenic species, notably the Mycobacterium tuberculosis complex (MTBC). Several lines of evidence attribute to PE and PE_PGRS genes critical roles in mycobacterial pathogenicity. To get more insight into the nature of these genes, we sought to address their evolutionary trajectories in the group of smooth tubercle bacilli (STB), the putative ancestor of the clonal MTBC.
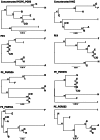
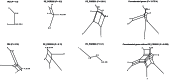
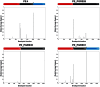
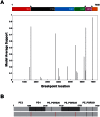
Methodology/principal findings: By focussing on six polymorphic STB PE/PE_PGRS genes, we demonstrate significant incongruence among single gene genealogies and detect strong signals of recombination using various approaches. Coalescent-based estimation of population recombination and mutation rates (ρ and θ, respectively) indicates that the two mechanisms are of roughly equal importance in generating diversity (ρ/θ = 1.457), a finding in a marked contrast to house keeping genes (HKG) whose evolution is chiefly brought about by mutation (ρ/θ = 0.012). In comparison to HKG, we found 15 times higher mean rate of nonsynonymous substitutions, with strong evidence of positive selection acting on PE_PGRS62 (dN/dS = 1.42), a gene that has previously been shown to be essential for mycobacterial survival in macrophages and granulomas. Imprint of positive selection operating on specific amino acid residues or along branches of PE_PGRS62 phylogenetic tree was further demonstrated using maximum likelihood- and covarion-based approaches, respectively. Strikingly, PE_PGR62 proved highly conserved in present-day MTBC strains.
Conclusions/significance: Overall the data indicate that, in STB, PE/PE_PGRS genes have undergone a strong diversification process that is speeded up by recombination, with evidence of positive selection. The finding that positive selection involved an essential PE_PGRS gene whose sequence appears to be driven to fixation in present-day MTBC strains lends further support to the critical role of PE/PE_PGRS genes in the evolution of mycobacterial pathogenicity.
Conflict of interest statement
Figures
References
-
- WHO (2011) Global Tuberculosis Control 2010. Geneva, Switzerland: World Health Organization.
-
- Canetti G (1970) Infection by atypical mycobacteria and antituberculous immunity. Lille Med 15: 280–282. - PubMed
-
- van Soolingen D, Hoogenboezem T, de Haas PE, Hermans PW, Koedam MA, et al. (1997) A novel pathogenic taxon of the Mycobacterium tuberculosis complex, Canetti: characterization of an exceptional isolate from Africa. Int J Syst Bacteriol 47: 1236–1245. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

