A model for the evolution of extremely fragmented macronuclei in ciliates
- PMID: 23705024
- PMCID: PMC3660376
- DOI: 10.1371/journal.pone.0064997
A model for the evolution of extremely fragmented macronuclei in ciliates
Abstract
While all ciliates possess nuclear dimorphism, several ciliates - like those in the classes Phyllopharyngea, Spirotrichea, and Armophorea - have an extreme macronuclear organization. Their extensively fragmented macronuclei contain upwards of 20,000 chromosomes, each with upwards of thousands of copies. These features have evolved independently on multiple occasions throughout ciliate evolutionary history, and currently no models explain these structures in an evolutionary context. In this paper, we propose that competition between two forces - the limitation and avoidance of chromosomal imbalances as a ciliate undergoes successive asexual divisions, and the costs of replicating massive genomes - is sufficient to explain this particular nuclear structure. We present a simulation of ciliate cell evolution under control of these forces, allowing certain features of the population to change over time. Over a wide range of parameters, we observe the repeated emergence of this unusual genomic organization found in nature. Although much remains to be understood about the evolution of macronuclear genome organization, our results show that the proposed model is a plausible explanation for the emergence of these extremely fragmented, highly polyploid genomes.
Conflict of interest statement
Figures
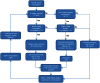
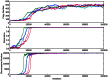
Similar articles
-
Macronuclear development in ciliates, with a focus on nuclear architecture.J Eukaryot Microbiol. 2022 Sep;69(5):e12898. doi: 10.1111/jeu.12898. Epub 2022 Mar 16. J Eukaryot Microbiol. 2022. PMID: 35178799 Free PMC article. Review.
-
Alternative processing of scrambled genes generates protein diversity in the ciliate Chilodonella uncinata.J Exp Zool B Mol Dev Evol. 2010 Sep 15;314(6):480-8. doi: 10.1002/jez.b.21354. J Exp Zool B Mol Dev Evol. 2010. PMID: 20700892 Free PMC article.
-
Variation in macronuclear genome content of three ciliates with extensive chromosomal fragmentation: a preliminary analysis.J Eukaryot Microbiol. 2007 May-Jun;54(3):242-6. doi: 10.1111/j.1550-7408.2007.00257.x. J Eukaryot Microbiol. 2007. PMID: 17552979
-
Evolution of amitosis of the ciliate macronucleus: gain of the capacity to divide.J Protozool. 1991 May-Jun;38(3):217-21. doi: 10.1111/j.1550-7408.1991.tb04431.x. J Protozool. 1991. PMID: 1908902 Review.
-
Evolution of germline-limited sequences in two populations of the ciliate Chilodonella uncinata.J Mol Evol. 2012 Apr;74(3-4):140-6. doi: 10.1007/s00239-012-9493-4. Epub 2012 Mar 13. J Mol Evol. 2012. PMID: 22411695
Cited by
-
Macronuclear development in ciliates, with a focus on nuclear architecture.J Eukaryot Microbiol. 2022 Sep;69(5):e12898. doi: 10.1111/jeu.12898. Epub 2022 Mar 16. J Eukaryot Microbiol. 2022. PMID: 35178799 Free PMC article. Review.
-
Insights into an Extensively Fragmented Eukaryotic Genome: De Novo Genome Sequencing of the Multinuclear Ciliate Uroleptopsis citrina.Genome Biol Evol. 2018 Mar 1;10(3):883-894. doi: 10.1093/gbe/evy055. Genome Biol Evol. 2018. PMID: 29608728 Free PMC article.
-
Programmed chromosome fragmentation in ciliated protozoa: multiple means to chromosome ends.Microbiol Mol Biol Rev. 2023 Dec 20;87(4):e0018422. doi: 10.1128/mmbr.00184-22. Epub 2023 Nov 27. Microbiol Mol Biol Rev. 2023. PMID: 38009915 Free PMC article. Review.
-
Structure of the germline genome of Tetrahymena thermophila and relationship to the massively rearranged somatic genome.Elife. 2016 Nov 28;5:e19090. doi: 10.7554/eLife.19090. Elife. 2016. PMID: 27892853 Free PMC article.
References
-
- Coyne RS, Chalker DL, Yao M (1996) Genome downsizing during ciliate development: Nuclear division of labor through chromosome restructuring. Annu Rev Genet 30 ((1)) 557–578. - PubMed
-
- Eisen JA, Coyne RS, Wu M, Wu D, Thiagarajan M, et al. (2006) Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol 4: e286 doi: 10.1371/journal.pbio.0040286. - DOI - PMC - PubMed
-
- Coyne RS, Hannick L, Shanmugam D, Hostetler JB, Brami D, et al. (2011) Comparative genomics of the pathogenic ciliate Ichthyophthirius multifiliis, its free- living relatives and a host species provide insights into adoption of a parasitic lifestyle and prospects for disease control. Genome Biol 12: R100. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

