Baseline survey of the anatomical microbial ecology of an important food plant: Solanum lycopersicum (tomato)
- PMID: 23705801
- PMCID: PMC3680157
- DOI: 10.1186/1471-2180-13-114
Baseline survey of the anatomical microbial ecology of an important food plant: Solanum lycopersicum (tomato)
Abstract
Background: Research to understand and control microbiological risks associated with the consumption of fresh fruits and vegetables has examined many environments in the farm to fork continuum. An important data gap however, that remains poorly studied is the baseline description of microflora that may be associated with plant anatomy either endemically or in response to environmental pressures. Specific anatomical niches of plants may contribute to persistence of human pathogens in agricultural environments in ways we have yet to describe. Tomatoes have been implicated in outbreaks of Salmonella at least 17 times during the years spanning 1990 to 2010. Our research seeks to provide a baseline description of the tomato microbiome and possibly identify whether or not there is something distinctive about tomatoes or their growing ecology that contributes to persistence of Salmonella in this important food crop.
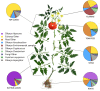
Results: DNA was recovered from washes of epiphytic surfaces of tomato anatomical organs; leaves, stems, roots, flowers and fruits of Solanum lycopersicum (BHN602), grown at a site in close proximity to commercial farms previously implicated in tomato-Salmonella outbreaks. DNA was amplified for targeted 16S and 18S rRNA genes and sheared for shotgun metagenomic sequencing. Amplicons and metagenomes were used to describe "native" bacterial microflora for diverse anatomical parts of Virginia-grown tomatoes.
Conclusions: Distinct groupings of microbial communities were associated with different tomato plant organs and a gradient of compositional similarity could be correlated to the distance of a given plant part from the soil. Unique bacterial phylotypes (at 95% identity) were associated with fruits and flowers of tomato plants. These include Microvirga, Pseudomonas, Sphingomonas, Brachybacterium, Rhizobiales, Paracocccus, Chryseomonas and Microbacterium. The most frequently observed bacterial taxa across aerial plant regions were Pseudomonas and Xanthomonas. Dominant fungal taxa that could be identified to genus with 18S amplicons included Hypocrea, Aureobasidium and Cryptococcus. No definitive presence of Salmonella could be confirmed in any of the plant samples, although 16S sequences suggested that closely related genera were present on leaves, fruits and roots.
Figures





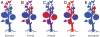
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

