Human milk metagenome: a functional capacity analysis
- PMID: 23705844
- PMCID: PMC3679945
- DOI: 10.1186/1471-2180-13-116
Human milk metagenome: a functional capacity analysis
Abstract
Background: Human milk contains a diverse population of bacteria that likely influences colonization of the infant gastrointestinal tract. Recent studies, however, have been limited to characterization of this microbial community by 16S rRNA analysis. In the present study, a metagenomic approach using Illumina sequencing of a pooled milk sample (ten donors) was employed to determine the genera of bacteria and the types of bacterial open reading frames in human milk that may influence bacterial establishment and stability in this primal food matrix. The human milk metagenome was also compared to that of breast-fed and formula-fed infants' feces (n = 5, each) and mothers' feces (n = 3) at the phylum level and at a functional level using open reading frame abundance. Additionally, immune-modulatory bacterial-DNA motifs were also searched for within human milk.
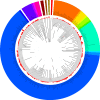
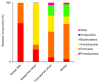
Results: The bacterial community in human milk contained over 360 prokaryotic genera, with sequences aligning predominantly to the phyla of Proteobacteria (65%) and Firmicutes (34%), and the genera of Pseudomonas (61.1%), Staphylococcus (33.4%) and Streptococcus (0.5%). From assembled human milk-derived contigs, 30,128 open reading frames were annotated and assigned to functional categories. When compared to the metagenome of infants' and mothers' feces, the human milk metagenome was less diverse at the phylum level, and contained more open reading frames associated with nitrogen metabolism, membrane transport and stress response (P < 0.05). The human milk metagenome also contained a similar occurrence of immune-modulatory DNA motifs to that of infants' and mothers' fecal metagenomes.
Conclusions: Our results further expand the complexity of the human milk metagenome and enforce the benefits of human milk ingestion on the microbial colonization of the infant gut and immunity. Discovery of immune-modulatory motifs in the metagenome of human milk indicates more exhaustive analyses of the functionality of the human milk metagenome are warranted.
Figures


References
-
- Kramer MS, Guo T, Platt RW, Sevkovskaya Z, Dzikovich I, Collet JP, Shapiro S, Chalmers B, Hodnett E, Vanilovich I, Mezen I, Ducruet T, Shishko G, Bogdanovich N. Infant growth and health outcomes associated with 3 compared with 6 mo of exclusive breastfeeding. Am J Clin Nutr. 2003;78:291–295. - PubMed
-
- Ladomenou F, Moschandreas J, Kafatos A, Tselentis Y, Galanakis E. Protective effect of exclusive breastfeeding against infections during infancy: a prospective study. Arch Dis Child. 2010;95:1004–1008. - PubMed
-
- Sangild PT, Siggers RH, Schmidt M, Elnif J, Bjornvad CR, Thymann T, Grondahl ML, Hansen AK, Jensen SK, Boye M, Moelbak L, Buddington RK, Westrom BR, Holst JJ, Burrin DG. Diet- and colonization-dependent intestinal dysfunction predisposes to necrotizing enterocolitis in preterm pigs. Gastroenterol. 2006;130:1776–1792. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

