The HIV-1 pandemic: does the selective sweep in chimpanzees mirror humankind's future?
- PMID: 23705941
- PMCID: PMC3667106
- DOI: 10.1186/1742-4690-10-53
The HIV-1 pandemic: does the selective sweep in chimpanzees mirror humankind's future?
Abstract
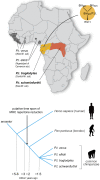
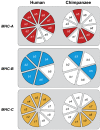
An HIV-1 infection progresses in most human individuals sooner or later into AIDS, a devastating disease that kills more than a million people worldwide on an annual basis. Nonetheless, certain HIV-1-infected persons appear to act as long-term non-progressors, and elite control is associated with the presence of particular MHC class I allotypes such as HLA-B*27 or -B*57. The HIV-1 pandemic in humans arose from the cross-species transmission of SIVcpz originating from chimpanzees. Chimpanzees, however, appear to be relatively resistant to developing AIDS after HIV-1/SIVcpz infection. Mounting evidence illustrates that, in the distant past, chimpanzees experienced a selective sweep resulting in a severe reduction of their MHC class I repertoire. This was most likely caused by an HIV-1/SIV-like retrovirus, suggesting that chimpanzees may have experienced long-lasting host-virus relationships with SIV-like viruses. Hence, if natural selection is allowed to follow its course, prospects for the human population may look grim, thus underscoring the desperate need for an effective vaccine.
Figures
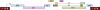



References
-
- Davey RT Jr, Bhat N, Yoder C, Chun TW, Metcalf JA, Dewar R, Natarajan V, Lempicki RA, Adelsberger JW, Miller KD. HIV-1 and T cell dynamics after interruption of highly active antiretroviral therapy (HAART) in patients with a history of sustained viral suppression. Proc Natl Acad Sci USA. 1999;96:15109–15114. doi: 10.1073/pnas.96.26.15109. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

