Molecular investigation of Torque teno sus virus in geographically distinct porcine breeding herds of Sichuan, China
- PMID: 23705989
- PMCID: PMC3679838
- DOI: 10.1186/1743-422X-10-161
Molecular investigation of Torque teno sus virus in geographically distinct porcine breeding herds of Sichuan, China
Abstract
Background: Torque teno sus virus (TTSuV), infecting domestic swine and wild boar, is a non-enveloped virus with a circular, single-stranded DNA genome. which has been classified into the genera Iotatorquevirus (TTSuV1) and Kappatorquevirus (TTSuV2) of the family Anelloviridae. A molecular study was conducted to detect evidence of a phylogenic relationship between these two porcine TTSuV genogroups from the sera of 244 infected pigs located in 21 subordinate prefectures and/or cities of Sichuan.
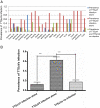
Results: Both genogroups of TTSuV were detected in pig sera collected from all 21 regions examined. Of the 244 samples, virus from either genogroup was detected in 203 (83.2%), while 44 animals (18.0%) were co-infected with viruses of both genogroups. Moreover, TTSuV2 (186/244, 76.2%) was more prevalent than TTSuV1 (61/244, 25%). There was statistically significant difference between the prevalence of genogroups 1 infection alone (9.4%, 23/244) and 2 alone (64.8%, 158/244), and between the prevalence of genogroups 2 (76.2%, 186/244) and both genogroups co-infection (18.0%, 44/244). The untranslated region of the swine TTSuV genome was found to be an adequate molecular marker of the virus for detection and surveillance. Phylogenetic analysis indicated that both genogroups 1 and 2 could be further divided into two subtypes, subtype a and b. TTSuV1 subtype b and the two TTSuV2 subtypes are more prevalent in Sichuan Province.
Conclusions: Our study presents detailed geographical evidence of TTSuV infection in China.
Figures


References
-
- Meng XJ. Emerging and Re-emerging swine viruses. Transbound Emerg Dis. 2012;59(S1):85–102. - PubMed
-
- Kekarainen T, Segales J. Torque teno Sus virus in pigs: an emerging pathogen? Transbound Emerg Dis. 2012;59(S1):103–108. - PubMed
-
- Leary TP, Erker JC, Chalmers ML, Desai SM, Mushahwar IK. Improved detection systems for TT virus reveal high prevalence in humans, non-human primates and farm animals. J Gen Virol. 1999;80(Pt 8):2115–2120. - PubMed
-
- Okamoto H, Takahashi M, Nishizawa T, Tawara A, Fukai K, Muramatsu U, Naito Y, Yoshikawa A. Genomic characterization of TT viruses (TTVs) in pigs, cats and dogs and their relatedness with species-specific TTVs in primates and tupaias. J Gen Virol. 2002;83:1291–1297. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

