The eEF2 kinase confers resistance to nutrient deprivation by blocking translation elongation
- PMID: 23706743
- PMCID: PMC4395874
- DOI: 10.1016/j.cell.2013.04.055
The eEF2 kinase confers resistance to nutrient deprivation by blocking translation elongation
Abstract
Metabolic adaptation is essential for cell survival during nutrient deprivation. We report that eukaryotic elongation factor 2 kinase (eEF2K), which is activated by AMP-kinase (AMPK), confers cell survival under acute nutrient depletion by blocking translation elongation. Tumor cells exploit this pathway to adapt to nutrient deprivation by reactivating the AMPK-eEF2K axis. Adaptation of transformed cells to nutrient withdrawal is severely compromised in cells lacking eEF2K. Moreover, eEF2K knockdown restored sensitivity to acute nutrient deprivation in highly resistant human tumor cell lines. In vivo, overexpression of eEF2K rendered murine tumors remarkably resistant to caloric restriction. Expression of eEF2K strongly correlated with overall survival in human medulloblastoma and glioblastoma multiforme. Finally, C. elegans strains deficient in efk-1, the eEF2K ortholog, were severely compromised in their response to nutrient depletion. Our data highlight a conserved role for eEF2K in protecting cells from nutrient deprivation and in conferring tumor cell adaptation to metabolic stress. PAPERCLIP:
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures
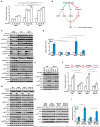


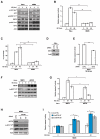
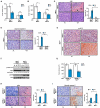


Comment in
-
Adaptation to starvation: translating a matter of life or death.Cancer Cell. 2013 Jun 10;23(6):713-5. doi: 10.1016/j.ccr.2013.05.012. Cancer Cell. 2013. PMID: 23763997
References
-
- Baugh LR, Demodena J, Sternberg PW. RNA Pol II accumulates at promoters of growth genes during developmental arrest. Science. 2009;324:92–94. - PubMed
-
- Braunstein S, Karpisheva K, Pola C, Goldberg J, Hochman T, Yee H, Cangiarella J, Arju R, Formenti SC, Schneider RJ. A hypoxia-controlled cap-dependent to cap-independent translation switch in breast cancer. Mol. Cell. 2007;28:501–512. - PubMed
-
- Buzzai M, Bauer DE, Jones RG, Deberardinis RJ, Hatzivassiliou G, Elstrom RL, Thompson CB. The glucose dependence of Akt-transformed cells can be reversed by pharmacologic activation of fatty acid beta-oxidation. Oncogene. 2005;24:4165–4173. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

