An aberrant transcription factor network essential for Wnt signaling and stem cell maintenance in glioblastoma
- PMID: 23707066
- PMCID: PMC3774301
- DOI: 10.1016/j.celrep.2013.04.021
An aberrant transcription factor network essential for Wnt signaling and stem cell maintenance in glioblastoma
Abstract
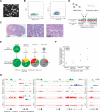
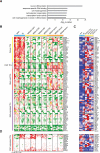
Glioblastoma (GBM) is thought to be driven by a subpopulation of cancer stem cells (CSCs) that self-renew and recapitulate tumor heterogeneity yet remain poorly understood. Here, we present a comparative analysis of chromatin state in GBM CSCs that reveals widespread activation of genes normally held in check by Polycomb repressors. These activated targets include a large set of developmental transcription factors (TFs) whose coordinated activation is unique to the CSCs. We demonstrate that a critical factor in the set, ASCL1, activates Wnt signaling by repressing the negative regulator DKK1. We show that ASCL1 is essential for the maintenance and in vivo tumorigenicity of GBM CSCs. Genome-wide binding profiles for ASCL1 and the Wnt effector LEF-1 provide mechanistic insight and suggest widespread interactions between the TF module and the signaling pathway. Our findings demonstrate regulatory connections among ASCL1, Wnt signaling, and collaborating TFs that are essential for the maintenance and tumorigenicity of GBM CSCs.
Copyright © 2013 The Authors. Published by Elsevier Inc. All rights reserved.
Figures



References
-
- Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. - PubMed
-
- Barde I, Salmon P, Trono D. Production and titration of lentiviral vectors. Curr Protoc Neurosci. 2010 Chapter 4, Unit 4 21. - PubMed
-
- Bell O, Tiwari VK, Thoma NH, Schubeler D. Determinants and dynamics of genome accessibility. Nat Rev Genet. 2011;12:554–564. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

