Partitioning RS domain phosphorylation in an SR protein through the CLK and SRPK protein kinases
- PMID: 23707382
- PMCID: PMC3860265
- DOI: 10.1016/j.jmb.2013.05.013
Partitioning RS domain phosphorylation in an SR protein through the CLK and SRPK protein kinases
Abstract
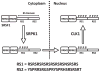
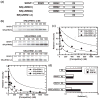
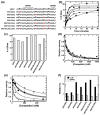
SR proteins are essential splicing factors whose biological function is regulated through phosphorylation of their C-terminal RS domains. Prior studies have shown that cytoplasmic-nuclear translocalization of the SR protein SRSF1 is regulated by multisite phosphorylation of a long Arg-Ser repeat in the N-terminus of the RS domain while subnuclear localization is controlled by phosphorylation of a shorter Arg-Ser repeat along with several Ser-Pro dipeptides in the C-terminus of the RS domain. To better understand how these two kinases partition Arg-Ser versus Ser-Pro specificities, we monitored the phosphorylation of SRSF1 by CLK1 and SRPK1. Although SRPK1 initially binds at the center of the RS domain phosphorylating in an orderly, N-terminal direction, CLK1 makes widespread contacts in the RS domain and generates multiple enzyme-substrate complexes that induce a random addition mechanism. While SRPK1 rapidly phosphorylates N-terminal serines, SRPK1 and CLK1 display similar activities toward Arg-Ser repeats in the C-terminus, suggesting that these kinases may not separate function in a strict linear manner along the RS domain. CLK1 induces a unique gel shift in SRSF1 that is not the result of enhanced Arg-Ser phosphorylation but rather is the direct result of the phosphorylation of several Ser-Pro dipeptides. These prolines are important for binding and phosphorylation of the SR protein by CLK1 but not for the SRPK1-dependent reaction. The data establish a new view of SR protein regulation in which SRPK1 and CLK1 partition activities based on Ser-Pro versus Arg-Ser placement rather than on N- and C-terminal preferences along the RS domain.
Keywords: BSA; CLK; Cdc2-like kinase; NIH; National Institutes of Health; RNA recognition motif; RRM; SR protein; SR-specific protein kinase; SRPK; bovine serum albumin; kinase; kinetics; phosphorylation; splicing.
Copyright © 2013 Elsevier Ltd. All rights reserved.
Figures





References
-
- Stojdl DF, Bell JC. SR protein kinases: the splice of life. Biochem Cell Biol. 1999;77:293–298. - PubMed
-
- Lai MC, Lin RI, Huang SY, Tsai CW, Tarn WY. A human importin-β family protein, transportin-SR2, interacts with the phosphorylated RS domain of SR proteins. J Biol Chem. 2000;275:7950–7957. - PubMed
-
- Mathew R, Hartmuth K, Mohlmann S, Urlaub H, Ficner R, Luhrmann R. Phosphorylation of human PRP28 by SRPK2 is required for integration of the U4/U6-U5 tri-snRNP into the spliceosome. Nat Struct Mol Biol. 2008;15:435–443. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

