Cellular and molecular players in adipose tissue inflammation in the development of obesity-induced insulin resistance
- PMID: 23707515
- PMCID: PMC3800253
- DOI: 10.1016/j.bbadis.2013.05.017
Cellular and molecular players in adipose tissue inflammation in the development of obesity-induced insulin resistance
Abstract
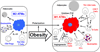
There is increasing evidence showing that inflammation is an important pathogenic mediator of the development of obesity-induced insulin resistance. It is now generally accepted that tissue-resident immune cells play a major role in the regulation of this obesity-induced inflammation. The roles that adipose tissue (AT)-resident immune cells play have been particularly extensively studied. AT contains most types of immune cells and obesity increases their numbers and activation levels, particularly in AT macrophages (ATMs). Other pro-inflammatory cells found in AT include neutrophils, Th1 CD4 T cells, CD8 T cells, B cells, DCs, and mast cells. However, AT also contains anti-inflammatory cells that counter the pro-inflammatory immune cells that are responsible for the obesity-induced inflammation in this tissue. These anti-inflammatory cells include regulatory CD4 T cells (Tregs), Th2 CD4 T cells, and eosinophils. Hence, AT inflammation is shaped by the regulation of pro- and anti-inflammatory immune cell homeostasis, and obesity skews this balance towards a more pro-inflammatory status. Recent genetic studies revealed several molecules that participate in the development of obesity-induced inflammation and insulin resistance. In this review, the cellular and molecular players that participate in the regulation of obesity-induced inflammation and insulin resistance are discussed, with particular attention being placed on the roles of the cellular players in these pathogeneses. This article is part of a Special Issue entitled: Modulation of Adipose Tissue in Health and Disease.
Keywords: ABCA; AIM; APC; ASC; AT; AT macrophage; ATM; ATP-binding cassette transporter; Adipose tissue; Adipose tissue immune cell; Apoptosis inhibitor of macrophage; BLT; BM; BMT; Bregs; C-C chemokine receptor type 2; C-X-C motif chemokine 5; C-X-C motif receptor 2; CCL5; CCR2; CLSs; CTLs; CX3C chemokine receptor 1; CX3CR1; CXCL5; CXCR2; Casp1; Caspase 1; Chemokine (C-C motif) ligand 5; DAMPs; DC; Damage-associated molecular pattern molecules; Dendritic cells; ECM; FABP; FFA; Free fatty acids; G-protein coupled receptor 120; GFP; GLUT4; GPR120; Glucose transporter type 4; HFD; HMGB1; IKKb; IR; IRS; Insulin receptor substrate; Insulin resistance; JNKs; Jun N-terminal kinases; KLF4; Krueppel-like factor 4; MAPK; MCP-1; MGL1; MHC; MPO; Major histocompatibility complex; NE; NFkB; NKT; NLS; NOD-like receptor family, pyrin domain containing 3; Natural Killer T cell; Nlrp3; Obesity-induced Inflammation; PBMC; PKC; PPAR; PRRs; Pattern recognition receptors; Protein kinase C; RAGs; RANTES; ROS; Reactive oxygen species; Recombination activating genes; Regulated on Activation Normal T cell Expressed and Secreted; SOCS1; SVC; Sorbin and SH3 domain-containing protein 1; Sorbs1; Suppressor of cytokine signaling 1; T cell receptors; T2D; TAMs; TCRs; TLR; TNF; TZDs; Toll-like receptors; Treg; Tumor necrosis factors; Type 2 Diabetes; adipose tissue; antigen-presenting cell; apoptotic speck protein containing a caspase recruitment domain; bone marrow; bone marrow transplantation; crown-like structures; cytotoxic T cells; double stranded DNA; dsDNA; extracellular matrix; fatty-acid-binding protein; green fluorescent protein; high fat diet; high-mobility group box 1; inhibitor of κB kinase-β; insulin receptor; leukotriene B4 receptor; macrophage galactose-type lectin 1; mitogen-activated protein kinase; monocyte chemotactic protein-1; myeloperoxidase; neutrophil elastase; nuclear factor kappa-light-chain-enhancer of activated B cells; nuclear localization sequence; peripheral blood mononuclear cell; peroxisome proliferator-activated receptors; regulator T cells; regulatory B cells; stromal vascular cell; thiazolidinediones; tumor-associated macrophages.
Copyright © 2013 Elsevier B.V. All rights reserved.
Figures

Similar articles
-
Genetic and Epigenetic Regulation of the Innate Immune Response to Gout.Immunol Invest. 2023 Apr;52(3):364-397. doi: 10.1080/08820139.2023.2168554. Epub 2023 Feb 6. Immunol Invest. 2023. PMID: 36745138 Review.
-
Reprint of Neutrophil cell surface receptors and their intracellular signal transduction pathways.Int Immunopharmacol. 2013 Dec;17(4):1185-97. doi: 10.1016/j.intimp.2013.11.010. Epub 2013 Nov 18. Int Immunopharmacol. 2013. PMID: 24263067 Review.
-
Neutrophil cell surface receptors and their intracellular signal transduction pathways.Int Immunopharmacol. 2013 Nov;17(3):638-50. doi: 10.1016/j.intimp.2013.06.034. Epub 2013 Aug 30. Int Immunopharmacol. 2013. PMID: 23994464 Free PMC article. Review.
-
Reappraisal of Adipose Tissue Inflammation in Obesity.Adv Exp Med Biol. 2024;1460:297-327. doi: 10.1007/978-3-031-63657-8_10. Adv Exp Med Biol. 2024. PMID: 39287856 Review.
-
Message Transmission Between Adipocyte and Macrophage in Obesity.Adv Exp Med Biol. 2024;1460:273-295. doi: 10.1007/978-3-031-63657-8_9. Adv Exp Med Biol. 2024. PMID: 39287855 Review.
Cited by
-
Stearic Acid and TNF-α Co-Operatively Potentiate MIP-1α Production in Monocytic Cells via MyD88 Independent TLR4/TBK/IRF3 Signaling Pathway.Biomedicines. 2020 Oct 9;8(10):403. doi: 10.3390/biomedicines8100403. Biomedicines. 2020. PMID: 33050324 Free PMC article.
-
Characterization and Treatment of Inflammation and Insulin Resistance in Obese Adipose Tissue.Diabetes Metab Syndr Obes. 2020 Oct 1;13:3449-3460. doi: 10.2147/DMSO.S271509. eCollection 2020. Diabetes Metab Syndr Obes. 2020. PMID: 33061505 Free PMC article. Review.
-
The role of macrophage subtypes and exosomes in immunomodulation.Cell Mol Biol Lett. 2022 Oct 3;27(1):83. doi: 10.1186/s11658-022-00384-y. Cell Mol Biol Lett. 2022. PMID: 36192691 Free PMC article. Review.
-
COVID-19 signalome: Pathways for SARS-CoV-2 infection and impact on COVID-19 associated comorbidity.Cell Signal. 2023 Jan;101:110495. doi: 10.1016/j.cellsig.2022.110495. Epub 2022 Oct 15. Cell Signal. 2023. PMID: 36252792 Free PMC article. Review.
-
Genetic inactivation of the LIGHT (TNFSF14) cytokine in mice restores glucose homeostasis and diminishes hepatic steatosis.Diabetologia. 2019 Nov;62(11):2143-2157. doi: 10.1007/s00125-019-4962-6. Epub 2019 Aug 6. Diabetologia. 2019. PMID: 31388695
References
-
- Kahn SE, Hull RL, Utzschneider KM. Mechanisms linking obesity to insulin resistance and type 2 diabetes. Nature. 2006;444:840–846. - PubMed
-
- Biddinger SB, Kahn CR. From mice to men: insights into the insulin resistance syndromes. Annu Rev Physiol. 2006;68:123–158. - PubMed
-
- White MF. Insulin signaling in health and disease. Science. 2003;302:1710–1711. - PubMed
-
- Pilch PF, Lee J. Insulin Receptor Family. In: Lennarz W, Lane MD, editors. Encyclopedia of Biological Chemistry. vol. 2. San Diego: Elsevier Science; 2004. pp. 436–440.
-
- Osborn O, Olefsky JM. The cellular and signaling networks linking the immune system and metabolism in disease. Nat Med. 2012;18:363–374. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

