Cloning, expression and purification of the SRCR domains of glycoprotein 340
- PMID: 23707657
- PMCID: PMC3727412
- DOI: 10.1016/j.pep.2013.05.003
Cloning, expression and purification of the SRCR domains of glycoprotein 340
Abstract
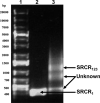
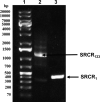
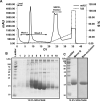
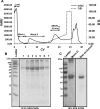
Glycoprotein 340 (gp340), an innate immunity molecule is secreted luminally by monolayered epithelia and associated glands within the human oral cavity. Gp340 contains 14 scavenger receptor cysteine rich (SRCR) domains, two CUB (C1r/C1s Uegf Bmp1) domains and one zona pellucida (ZP) domain. Oral streptococci are known to adhere to the tooth immobilized gp340 via its surface protein Antigen I/II (AgI/II), which is considered to be the critical first step in pathogenesis that eventually results in colonization and infection. In order to decipher the interactions between gp340's domains and oral streptococcal AgI/II domains, we undertook to express human gp340's first SRCR domain (SRCR1) and the first three tandem SRCR domains (SRCR123) in Drosophila S2 cells. While our initial attempts with human codons did not produce optimal results, codon-optimization for expression in Drosophila S2 cells and usage of inducible/secretory Drosophila expression system (DES) pMT/BiP/V5-HisA vector greatly enhanced the expression of the SRCR domains. Here we report the successful cloning, expression, and purification of the SRCR domains of gp340. Recognition of expressed SRCRs by the conformational dependent gp340 antibody indicate that these domains are appropriately folded and furthermore, surface plasmon resonance studies confirmed functional adherence of the SRCR domains to AgI/II.
Keywords: DMBT1; SAG; SRCR; Streptococcus mutans; gp340.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures



References
-
- Almstah IA, Wikstrom M, Stenberg I, Jakobsson A, Fagerberg-Mohlin B. Oral microbiota associated with hyposalivation of different origins. Oral Microbiol Immunol. 2003;18:1–8. - PubMed
-
- Moore WE, Moore LV. The bacteria of periodontal diseases. Periodontol 2000. 1994;5:66–77. - PubMed
-
- Hajishengallis G, Koga T, Russell MW. Affinity and specificity of the interactions between Streptococcus mutans antigen I/II and salivary components. J Dent Res. 1994;73:1493–1502. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

