The banana genome hub
- PMID: 23707967
- PMCID: PMC3662865
- DOI: 10.1093/database/bat035
The banana genome hub
Abstract
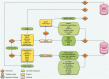
Banana is one of the world's favorite fruits and one of the most important crops for developing countries. The banana reference genome sequence (Musa acuminata) was recently released. Given the taxonomic position of Musa, the completed genomic sequence has particular comparative value to provide fresh insights about the evolution of the monocotyledons. The study of the banana genome has been enhanced by a number of tools and resources that allows harnessing its sequence. First, we set up essential tools such as a Community Annotation System, phylogenomics resources and metabolic pathways. Then, to support post-genomic efforts, we improved banana existing systems (e.g. web front end, query builder), we integrated available Musa data into generic systems (e.g. markers and genetic maps, synteny blocks), we have made interoperable with the banana hub, other existing systems containing Musa data (e.g. transcriptomics, rice reference genome, workflow manager) and finally, we generated new results from sequence analyses (e.g. SNP and polymorphism analysis). Several uses cases illustrate how the Banana Genome Hub can be used to study gene families. Overall, with this collaborative effort, we discuss the importance of the interoperability toward data integration between existing information systems. Database URL: http://banana-genome.cirad.fr/
Figures




Similar articles
-
Insights into the Musa genome: syntenic relationships to rice and between Musa species.BMC Genomics. 2008 Jan 30;9:58. doi: 10.1186/1471-2164-9-58. BMC Genomics. 2008. PMID: 18234080 Free PMC article.
-
The banana (Musa acuminata) genome and the evolution of monocotyledonous plants.Nature. 2012 Aug 9;488(7410):213-7. doi: 10.1038/nature11241. Nature. 2012. PMID: 22801500
-
Repetitive part of the banana (Musa acuminata) genome investigated by low-depth 454 sequencing.BMC Plant Biol. 2010 Sep 16;10:204. doi: 10.1186/1471-2229-10-204. BMC Plant Biol. 2010. PMID: 20846365 Free PMC article.
-
Expansion of banana (Musa acuminata) gene families involved in ethylene biosynthesis and signalling after lineage-specific whole-genome duplications.New Phytol. 2014 May;202(3):986-1000. doi: 10.1111/nph.12710. Epub 2014 Feb 7. New Phytol. 2014. PMID: 24716518
-
Discovery of new enzymatic functions and metabolic pathways using genomic enzymology web tools.Curr Opin Biotechnol. 2021 Jun;69:77-90. doi: 10.1016/j.copbio.2020.12.004. Epub 2021 Jan 5. Curr Opin Biotechnol. 2021. PMID: 33418450 Free PMC article. Review.
Cited by
-
Musa species in mainland Southeast Asia: From wild to domesticate.PLoS One. 2024 Oct 2;19(10):e0307592. doi: 10.1371/journal.pone.0307592. eCollection 2024. PLoS One. 2024. PMID: 39356650 Free PMC article.
-
Genome-wide identification and expression profiling of WUSCHEL-related homeobox (WOX) genes confer their roles in somatic embryogenesis, growth and abiotic stresses in banana.3 Biotech. 2022 Nov;12(11):321. doi: 10.1007/s13205-022-03387-w. Epub 2022 Oct 12. 3 Biotech. 2022. PMID: 36276441 Free PMC article.
-
The genetic mechanisms underlying the convergent evolution of pollination syndromes in the Neotropical radiation of Costus L.Front Plant Sci. 2022 Sep 8;13:874322. doi: 10.3389/fpls.2022.874322. eCollection 2022. Front Plant Sci. 2022. PMID: 36161003 Free PMC article.
-
From fruit growth to ripening in plantain: a careful balance between carbohydrate synthesis and breakdown.J Exp Bot. 2022 Aug 11;73(14):4832-4849. doi: 10.1093/jxb/erac187. J Exp Bot. 2022. PMID: 35512676 Free PMC article.
-
GREENC: a Wiki-based database of plant lncRNAs.Nucleic Acids Res. 2016 Jan 4;44(D1):D1161-6. doi: 10.1093/nar/gkv1215. Epub 2015 Nov 17. Nucleic Acids Res. 2016. PMID: 26578586 Free PMC article.
References
-
- D’Hont A, Denoeud F, Aury JM, et al. The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature. 2012;488:213–217. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

