A pan-specific antibody for direct detection of protein histidine phosphorylation
- PMID: 23708076
- PMCID: PMC3686892
- DOI: 10.1038/nchembio.1259
A pan-specific antibody for direct detection of protein histidine phosphorylation
Abstract
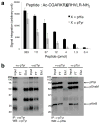
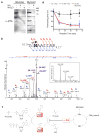
Despite its importance in central metabolism and bacterial cell signaling, protein histidine phosphorylation has remained elusive with respect to its extent and functional roles in biological systems because of the lack of adequate research tools. We report the development of the first pan-phosphohistidine (pHis) antibody using a stable pHis mimetic as the hapten. This antibody was successfully used in ELISA, western blotting, dot blot assays and immunoprecipitation and in detection and identification of histidine-phosphorylated proteins from native cell lysates when coupled with MS analysis. We also observed that the amount of protein pHis in Escherichia coli lysates depends on carbon source and nitrogen availability in the growth medium. In particular, we found that the amount of pHis on phosphoenolpyruvate synthase (PpsA) is sensitive to nitrogen availability in vivo and that α-ketoglutarate inhibits phosphotransfer from phosphorylated PpsA to pyruvate. We expect this antibody to open opportunities for investigating other pHis proteins and their functions.
Conflict of interest statement
J.-M.K. and T.W.M. are co-inventors of a patent application on stable phosphohistidine analogs.
Figures



Comment in
-
Post-translational modifications: Panning for phosphohistidine.Nat Chem Biol. 2013 Jul;9(7):411-2. doi: 10.1038/nchembio.1273. Epub 2013 May 26. Nat Chem Biol. 2013. PMID: 23708075 No abstract available.
-
Proteomics: Matching marks with mechanisms.Nat Methods. 2013 Jul;10(7):604-5. doi: 10.1038/nmeth.2546. Nat Methods. 2013. PMID: 23967483 No abstract available.
References
-
- Walsh CT. posttranslational modification of proteins: expanding nature’s inventory. Roberts and Company Publishers; Greenwood Village: 2006. Protein phosphorylation by protein kinases; pp. 35–80.
-
- Matthews HR. Protein kinases and phosphatases that act on histidine, lysine, or arginine residues in eukaryotic proteins: a possible regulator of the mitogen-activated protein kinase cascade. Pharmacol Therapeut. 1995;67:323–350. - PubMed
-
- Attwood PV, Besant PG, Piggott MJ. Focus on phosphoaspartate and phosphoglutamate. Amino Acids. 2010;40:1035–1051. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

