Recombination-induced tag exchange (RITE) cassette series to monitor protein dynamics in Saccharomyces cerevisiae
- PMID: 23708297
- PMCID: PMC3737166
- DOI: 10.1534/g3.113.006213
Recombination-induced tag exchange (RITE) cassette series to monitor protein dynamics in Saccharomyces cerevisiae
Abstract
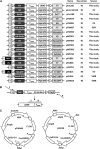
Proteins are not static entities. They are highly mobile, and their steady-state levels are achieved by a balance between ongoing synthesis and degradation. The dynamic properties of a protein can have important consequences for its function. For example, when a protein is degraded and replaced by a newly synthesized one, posttranslational modifications are lost and need to be reincorporated in the new molecules. Protein stability and mobility are also relevant for the duplication of macromolecular structures or organelles, which involves coordination of protein inheritance with the synthesis and assembly of newly synthesized proteins. To measure protein dynamics, we recently developed a genetic pulse-chase assay called recombination-induced tag exchange (RITE). RITE has been successfully used in Saccharomyces cerevisiae to measure turnover and inheritance of histone proteins, to study changes in posttranslational modifications on aging proteins, and to visualize the spatiotemporal inheritance of protein complexes and organelles in dividing cells. Here we describe a series of successful RITE cassettes that are designed for biochemical analyses, genomics studies, as well as single cell fluorescence applications. Importantly, the genetic nature and the stability of the tag switch offer the unique possibility to combine RITE with high-throughput screening for protein dynamics mutants and mechanisms. The RITE cassettes are widely applicable, modular by design, and can therefore be easily adapted for use in other cell types or organisms.
Keywords: epitope tag; protein inheritance; protein turnover; pulse-chase.
Figures



References
-
- Albert I., Mavrich T. N., Tomsho L. P., Qi J., Zanton S. J., et al. , 2007. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature 446: 572–576 - PubMed
-
- Brachmann C. B., Davies A., Cost G. J., Caputo E., Li J., et al. , 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14: 115–132 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
