DNA ejection from an archaeal virus--a single-molecule approach
- PMID: 23708366
- PMCID: PMC3660630
- DOI: 10.1016/j.bpj.2013.03.061
DNA ejection from an archaeal virus--a single-molecule approach
Abstract
The translocation of genetic material from the viral capsid to the cell is an essential part of the viral infection process. Whether the energetics of this process is driven by the energy stored within the confined nucleic acid or cellular processes pull the genome into the cell has been the subject of discussion. However, in vitro studies of genome ejection have been limited to a few head-tailed bacteriophages with a double-stranded DNA genome. Here we describe a DNA release system that operates in an archaeal virus. This virus infects an archaeon Haloarcula hispanica that was isolated from a hypersaline environment. The DNA-ejection velocity of His1, determined by single-molecule experiments, is comparable to that of bacterial viruses. We found that the ejection process is modulated by the external osmotic pressure (polyethylene glycol (PEG)) and by increased ion (Mg(2+) and Na(+)) concentration. The observed ejection was unidirectional, randomly paused, and incomplete, which suggests that cellular processes are required to complete the DNA transfer.
Copyright © 2013 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures


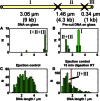

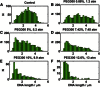
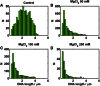
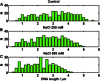
Comment in
-
Physical evolution of pressure-driven viral infection.Biophys J. 2013 May 21;104(10):2113-4. doi: 10.1016/j.bpj.2013.03.062. Biophys J. 2013. PMID: 23708348 Free PMC article. No abstract available.
References
-
- Suttle C.A. Marine viruses—major players in the global ecosystem. Nat. Rev. Microbiol. 2007;5:801–812. - PubMed
-
- Yang X., Yang H., Zhao G.P. Infectious disease in the genomic era. Annu. Rev. Genomics Hum. Genet. 2008;9:21–48. - PubMed
-
- Pina M., Bize A., Prangishvili D. The archeoviruses. FEMS Microbiol. Rev. 2011;35:1035–1054. - PubMed
-
- Atanasova N.S., Roine E., Oksanen H.M. Global network of specific virus-host interactions in hypersaline environments. Environ. Microbiol. 2012;14:426–440. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

