MicroRNA target site identification by integrating sequence and binding information
- PMID: 23708386
- PMCID: PMC3818907
- DOI: 10.1038/nmeth.2489
MicroRNA target site identification by integrating sequence and binding information
Abstract
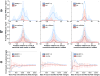
High-throughput sequencing has opened numerous possibilities for the identification of regulatory RNA-binding events. Cross-linking and immunoprecipitation of Argonaute proteins can pinpoint a microRNA (miRNA) target site within tens of bases but leaves the identity of the miRNA unresolved. A flexible computational framework, microMUMMIE, integrates sequence with cross-linking features and reliably identifies the miRNA family involved in each binding event. It considerably outperforms sequence-only approaches and quantifies the prevalence of noncanonical binding modes.
Figures

References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

