Electron microscopy structure of human APC/C(CDH1)-EMI1 reveals multimodal mechanism of E3 ligase shutdown
- PMID: 23708605
- PMCID: PMC3742808
- DOI: 10.1038/nsmb.2593
Electron microscopy structure of human APC/C(CDH1)-EMI1 reveals multimodal mechanism of E3 ligase shutdown
Abstract
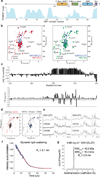
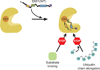
The anaphase-promoting complex/cyclosome (APC/C) is a ~1.5-MDa multiprotein E3 ligase enzyme that regulates cell division by promoting timely ubiquitin-mediated proteolysis of key cell-cycle regulatory proteins. Inhibition of human APC/C(CDH1) during interphase by early mitotic inhibitor 1 (EMI1) is essential for accurate coordination of DNA synthesis and mitosis. Here, we report a hybrid structural approach involving NMR, electron microscopy and enzymology, which reveal that EMI1's 143-residue C-terminal domain inhibits multiple APC/C(CDH1) functions. The intrinsically disordered D-box, linker and tail elements, together with a structured zinc-binding domain, bind distinct regions of APC/C(CDH1) to synergistically both block the substrate-binding site and inhibit ubiquitin-chain elongation. The functional importance of intrinsic structural disorder is explained by enabling a small inhibitory domain to bind multiple sites to shut down various functions of a 'molecular machine' nearly 100 times its size.
Figures





Comment in
-
EMI1, a three-in-one ubiquitylation inhibitor.Nat Struct Mol Biol. 2013 Jul;20(7):773-4. doi: 10.1038/nsmb.2626. Nat Struct Mol Biol. 2013. PMID: 23984442
Similar articles
-
Atomic structure of the APC/C and its mechanism of protein ubiquitination.Nature. 2015 Jun 25;522(7557):450-454. doi: 10.1038/nature14471. Epub 2015 Jun 15. Nature. 2015. PMID: 26083744 Free PMC article.
-
Emi1 stably binds and inhibits the anaphase-promoting complex/cyclosome as a pseudosubstrate inhibitor.Genes Dev. 2006 Sep 1;20(17):2410-20. doi: 10.1101/gad.1454006. Epub 2006 Aug 18. Genes Dev. 2006. PMID: 16921029 Free PMC article.
-
Structures of APC/C(Cdh1) with substrates identify Cdh1 and Apc10 as the D-box co-receptor.Nature. 2011 Feb 10;470(7333):274-8. doi: 10.1038/nature09625. Epub 2010 Nov 24. Nature. 2011. PMID: 21107322 Free PMC article.
-
Insights into the anaphase-promoting complex: a molecular machine that regulates mitosis.Curr Opin Struct Biol. 2014 Dec;29:1-9. doi: 10.1016/j.sbi.2014.08.003. Epub 2014 Aug 24. Curr Opin Struct Biol. 2014. PMID: 25174288 Review.
-
Structure, function and mechanism of the anaphase promoting complex (APC/C).Q Rev Biophys. 2011 May;44(2):153-90. doi: 10.1017/S0033583510000259. Epub 2010 Nov 22. Q Rev Biophys. 2011. PMID: 21092369 Review.
Cited by
-
ProteoPlex: stability optimization of macromolecular complexes by sparse-matrix screening of chemical space.Nat Methods. 2015 Sep;12(9):859-65. doi: 10.1038/nmeth.3493. Epub 2015 Aug 3. Nat Methods. 2015. PMID: 26237227 Free PMC article.
-
The pseudosubstrate inhibitor Acm1 inhibits the anaphase-promoting complex/cyclosome by combining high-affinity activator binding with disruption of Doc1/Apc10 function.J Biol Chem. 2019 Nov 15;294(46):17249-17261. doi: 10.1074/jbc.RA119.009468. Epub 2019 Sep 27. J Biol Chem. 2019. PMID: 31562243 Free PMC article.
-
Digested disorder: Quarterly intrinsic disorder digest (July-August-September, 2013).Intrinsically Disord Proteins. 2014 May 19;2(1):e27833. doi: 10.4161/idp.27833. eCollection 2014. Intrinsically Disord Proteins. 2014. PMID: 28232877 Free PMC article. Review.
-
De Novo-Designed APC/C Inhibitors Provide a Rationale for Targeting RING-Type E3 Ubiquitin Ligases.J Med Chem. 2025 Jun 12;68(11):11468-11483. doi: 10.1021/acs.jmedchem.5c00416. Epub 2025 May 21. J Med Chem. 2025. PMID: 40397069 Free PMC article.
-
Development of D-box peptides to inhibit the anaphase-promoting complex/cyclosome.Elife. 2025 Sep 1;14:RP104238. doi: 10.7554/eLife.104238. Elife. 2025. PMID: 40888475 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

