Phosphorylation of Nanog is essential to regulate Bmi1 and promote tumorigenesis
- PMID: 23708658
- PMCID: PMC3912208
- DOI: 10.1038/onc.2013.173
Phosphorylation of Nanog is essential to regulate Bmi1 and promote tumorigenesis
Abstract
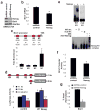
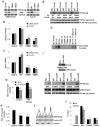
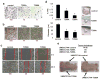
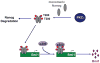
Emerging evidence indicates that Nanog is intimately involved in tumorigenesis, in part, through regulation of the cancer-initiating cell (CIC) population. However, the regulation and role of Nanog in tumorigenesis are still poorly understood. In this study, human Nanog was identified to be phosphorylated by human protein kinase Cɛ at multiple residues, including T200 and T280. Our work indicated that phosphorylation at T200 and T280 modulates Nanog function through several regulatory mechanisms. Results with phosphorylation-insensitive and phosphorylation-mimetic mutant Nanog revealed that phosphorylation at T200 and T280 enhance Nanog protein stability. Moreover, phosphorylation-insensitive T200A and T280A mutant Nanog had a dominant-negative function to inhibit endogenous Nanog transcriptional activity. Inactivation of Nanog was due to impaired homodimerization, DNA binding, promoter occupancy and p300, a transcriptional co-activator, recruitment resulting in a defect in target gene-promoter activation. Ectopic expression of phosphorylation-insensitive T200A or T280A mutant Nanog reduced cell proliferation, colony formation, invasion, migration and the CIC population in head and neck squamous cell carcinoma (HNSCC) cells. The in vivo cancer-initiating ability was severely compromised in HNSCC cells expressing phosphorylation-insensitive T200A or T280A mutant Nanog; 87.5% (14/16), 12.5% (1/8), and 0% (0/8) for control, T200A, and T280A, respectively. Nanog occupied the Bmi1 promoter to directly transactivate and regulate Bmi1. Genetic ablation and rescue experiments demonstrated that Bmi1 is a critical downstream signaling node for the pleiotropic, pro-oncogenic effects of Nanog. Taken together, our study revealed, for the first time, that post-translational phosphorylation of Nanog is essential to regulate Bmi1 and promote tumorigenesis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000 Apr;24(4):372–6. - PubMed
-
- Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, et al. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003 May 30;113(5):631–42. - PubMed
-
- Masui S, Nakatake Y, Toyooka Y, Shimosato D, Yagi R, Takahashi K, et al. Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat Cell Biol. 2007 Jun;9(6):625–35. - PubMed
-
- Torres J, Watt FM. Nanog maintains pluripotency of mouse embryonic stem cells by inhibiting NFkappaB and cooperating with Stat3. Nat Cell Biol. 2008 Feb;10(2):194–201. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

