Ubiquitin-dependent recruitment of the Bloom syndrome helicase upon replication stress is required to suppress homologous recombination
- PMID: 23708797
- PMCID: PMC3680739
- DOI: 10.1038/emboj.2013.117
Ubiquitin-dependent recruitment of the Bloom syndrome helicase upon replication stress is required to suppress homologous recombination
Abstract
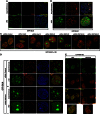
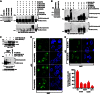
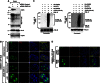
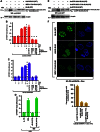
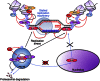
Limiting the levels of homologous recombination (HR) that occur at sites of DNA damage is a major role of BLM helicase. However, very little is known about the mechanisms dictating its relocalization to these sites. Here, we demonstrate that the ubiquitin/SUMO-dependent DNA damage response (UbS-DDR), controlled by the E3 ligases RNF8/RNF168, triggers BLM recruitment to sites of replication fork stalling via ubiquitylation in the N-terminal region of BLM and subsequent BLM binding to the ubiquitin-interacting motifs of RAP80. Furthermore, we show that this mechanism of BLM relocalization is essential for BLM's ability to suppress excessive/uncontrolled HR at stalled replication forks. Unexpectedly, we also uncovered a requirement for RNF8-dependent ubiquitylation of BLM and PML for maintaining the integrity of PML-associated nuclear bodies and as a consequence the localization of BLM to these structures. Lastly, we identified a novel role for RAP80 in preventing proteasomal degradation of BLM in unstressed cells. Taken together, these data highlight an important biochemical link between the UbS-DDR and BLM-dependent pathways involved in maintaining genome stability.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

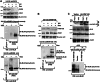

References
-
- Bekker-Jensen S, Rendtlew Danielsen J, Fugger K, Gromova I, Nerstedt A, Lukas C, Bartek J, Lukas J, Mailand N (2010) HERC2 coordinates ubiquitin-dependent assembly of DNA repair factors on damaged chromosomes. Nat Cell Biol 12: 80–86,sup pp 1–12 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

