The linear ubiquitin-specific deubiquitinase gumby regulates angiogenesis
- PMID: 23708998
- PMCID: PMC4931916
- DOI: 10.1038/nature12296
The linear ubiquitin-specific deubiquitinase gumby regulates angiogenesis
Abstract
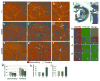
A complex interaction of signalling events, including the Wnt pathway, regulates sprouting of blood vessels from pre-existing vasculature during angiogenesis. Here we show that two distinct mutations in the (uro)chordate-specific gumby (also called Fam105b) gene cause an embryonic angiogenic phenotype in gumby mice. Gumby interacts with disheveled 2 (DVL2), is expressed in canonical Wnt-responsive endothelial cells and encodes an ovarian tumour domain class of deubiquitinase that specifically cleaves linear ubiquitin linkages. A crystal structure of gumby in complex with linear diubiquitin reveals how the identified mutations adversely affect substrate binding and catalytic function in line with the severity of their angiogenic phenotypes. Gumby interacts with HOIP (also called RNF31), a key component of the linear ubiquitin assembly complex, and decreases linear ubiquitination and activation of NF-κB-dependent transcription. This work provides support for the biological importance of linear (de)ubiquitination in angiogenesis, craniofacial and neural development and in modulating Wnt signalling.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



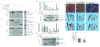
Comment in
-
Post-translational modifications: Breaking linear chains.Nat Rev Mol Cell Biol. 2013 Jul;14(7):402-3. doi: 10.1038/nrm3612. Epub 2013 Jun 19. Nat Rev Mol Cell Biol. 2013. PMID: 23778969 No abstract available.
References
-
- Risau W. Mechanisms of angiogenesis. Nature. 1997;386(6626):671–674. - PubMed
-
- Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. - PubMed
-
- Zerlin M, Julius MA, Kitajewski J. Wnt/Frizzled signaling in angiogenesis. Angiogenesis. 2008;11(1):63–69. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

