A defined methodology for reliable quantification of Western blot data
- PMID: 23709336
- PMCID: PMC3840294
- DOI: 10.1007/s12033-013-9672-6
A defined methodology for reliable quantification of Western blot data
Abstract
Chemiluminescent western blotting has been in common practice for over three decades, but its use as a quantitative method for measuring the relative expression of the target proteins is still debatable. This is mainly due to the various steps, techniques, reagents, and detection methods that are used to obtain the associated data. In order to have confidence in densitometric data from western blots, researchers should be able to demonstrate statistically significant fold differences in protein expression. This entails a necessary evolution of the procedures, controls, and the analysis methods. We describe a methodology to obtain reliable quantitative data from chemiluminescent western blots using standardization procedures coupled with the updated reagents and detection methods.
Conflict of interest statement
The authors declare no Conflict of Interest
Figures
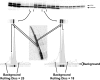

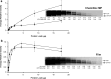




References
-
- Bio-Rad Laboratories (2011) Protein Blotting Guide, Interactive PDF. Review B, Bulletin 2895.
-
- Editorial A picture worth a thousand words (of explanation) Nature Methods. 2006;3(4):237. doi: 10.1038/nmeth0406-237. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

