Development of biotin-prototrophic and -hyperauxotrophic Corynebacterium glutamicum strains
- PMID: 23709504
- PMCID: PMC3719520
- DOI: 10.1128/AEM.00828-13
Development of biotin-prototrophic and -hyperauxotrophic Corynebacterium glutamicum strains
Abstract
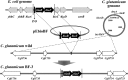
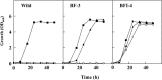
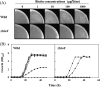
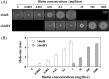
To develop the infrastructure for biotin production through naturally biotin-auxotrophic Corynebacterium glutamicum, we attempted to engineer the organism into a biotin prototroph and a biotin hyperauxotroph. To confer biotin prototrophy on the organism, the cotranscribed bioBF genes of Escherichia coli were introduced into the C. glutamicum genome, which originally lacked the bioF gene. The resulting strain still required biotin for growth, but it could be replaced by exogenous pimelic acid, a source of the biotin precursor pimelate thioester linked to either coenzyme A (CoA) or acyl carrier protein (ACP). To bridge the gap between the pimelate thioester and its dedicated precursor acyl-CoA (or -ACP), the bioI gene of Bacillus subtilis, which encoded a P450 protein that cleaves a carbon-carbon bond of an acyl-ACP to generate pimeloyl-ACP, was further expressed in the engineered strain by using a plasmid system. This resulted in a biotin prototroph that is capable of the de novo synthesis of biotin. On the other hand, the bioY gene responsible for biotin uptake was disrupted in wild-type C. glutamicum. Whereas the wild-type strain required approximately 1 μg of biotin per liter for normal growth, the bioY disruptant (ΔbioY) required approximately 1 mg of biotin per liter, almost 3 orders of magnitude higher than the wild-type level. The ΔbioY strain showed a similar high requirement for the precursor dethiobiotin, a substrate for bioB-encoded biotin synthase. To eliminate the dependency on dethiobiotin, the bioB gene was further disrupted in both the wild-type strain and the ΔbioY strain. By selectively using the resulting two strains (ΔbioB and ΔbioBY) as indicator strains, we developed a practical biotin bioassay system that can quantify biotin in the seven-digit range, from approximately 0.1 μg to 1 g per liter. This bioassay proved that the engineered biotin prototroph of C. glutamicum produced biotin directly from glucose, albeit at a marginally detectable level (approximately 0.3 μg per liter).
Figures


References
-
- Knowles JR. 1989. The mechanism of biotin-dependent enzymes. Annu. Rev. Biochem. 58:195–221 - PubMed
-
- Jitrapakdee S, Wallace JC. 2003. The biotin enzyme family: conserved structural motifs and domain rearrangements. Curr. Protein Pept. Sci. 4:217–229 - PubMed
-
- Shaw N, Lehner B, Fuhrmann M, Kulla H, Brass J, Birch O, Tinschert A, Venetz D, Venetz V, Sanchez JC, Tonella L, Hochstrasser D. 1999. Biotin production under limiting growth conditions by Agrobacterium/Rhizobium HK4 transformed with a modified Escherichia coli bio operon. J. Ind. Microbiol. Biotechnol. 22:590–599 - PubMed
-
- Streit WR, Entcheva P. 2003. Biotin in microbes, the genes involved in its biosynthesis, its biochemical role and perspectives for biotechnological production. Appl. Microbiol. Biotechnol. 61:21–31 - PubMed
-
- Goldberg MW, Sternbach LH. November 1949. Synthesis of biotin. US patent 2,489,232
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

