Complete genomes of two dipteran-associated spiroplasmas provided insights into the origin, dynamics, and impacts of viral invasion in spiroplasma
- PMID: 23711669
- PMCID: PMC3698928
- DOI: 10.1093/gbe/evt084
Complete genomes of two dipteran-associated spiroplasmas provided insights into the origin, dynamics, and impacts of viral invasion in spiroplasma
Abstract
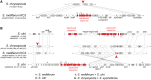
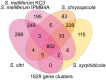
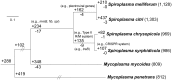
Spiroplasma is a genus of wall-less, low-GC, Gram-positive bacteria with helical morphology. As commensals or pathogens of plants, insects, ticks, or crustaceans, they are closely related with mycoplasmas and form a monophyletic group (Spiroplasma-Entomoplasmataceae-Mycoides) with Mycoplasma mycoides and its relatives. In this study, we report the complete genome sequences of Spiroplasma chrysopicola and S. syrphidicola from the Chrysopicola clade. These species form the sister group to the Citri clade, which includes several well-known pathogenic spiroplasmas. Surprisingly, these two newly available genomes from the Chrysopicola clade contain no plectroviral genes, which were found to be highly repetitive in the previously sequenced genomes from the Citri clade. Based on the genome alignment and patterns of GC-skew, these two Chrysopicola genomes appear to be relatively stable, rather than being highly rearranged as those from the Citri clade. Phylogenetic analyses suggest that the susceptibility to plectroviral invasion probably originated in the common ancestor of the Citri clade or one of its subclades. This susceptibility may be attributed to the absence of antiviral systems found in the Chrysopicola clade. Using the virus-free genomes of the Chrysopicola clade as references, we inferred the putative viral integration sites in the Citri genomes. Comparisons of syntenic regions suggest that the extensive viral invasion in the Citri clade promoted genome rearrangements and expansions. More importantly, the viral invasion may have facilitated horizontal gene transfers that contributed to adaptation in the Citri clade.
Keywords: Citri–Chrysopicola–Mirum clade; Mollicutes; Spiroplasma chrysopicola; Spiroplasma syrphidicola; plectrovirus; viral insertion.
Figures



References
-
- Alexeev D, et al. Application of Spiroplasma melliferum proteogenomic profiling for the discovery of virulence factors and pathogenicity mechanisms in host-associated spiroplasmas. J Proteome Res. 2012;11:224–236. - PubMed
-
- André A, Maccheroni W, Doignon F, Garnier M, Renaudin J. Glucose and trehalose PTS permeases of Spiroplasma citri probably share a single IIA domain, enabling the spiroplasma to adapt quickly to carbohydrate changes in its environment. Microbiology. 2003;149:2687–2696. - PubMed
-
- Bai X, Hogenhout SA. A genome sequence survey of the mollicute corn stunt spiroplasma Spiroplasma kunkelii. FEMS Microbiol Lett. 2002;210:7–17. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

