Rapid assessment of RNAi-mediated protein depletion by selected reaction monitoring mass spectrometry
- PMID: 23713831
- PMCID: PMC3767422
- DOI: 10.1021/pr400067k
Rapid assessment of RNAi-mediated protein depletion by selected reaction monitoring mass spectrometry
Abstract
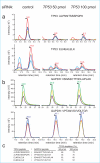
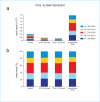
We describe the use of a targeted proteomics approach, selected reaction monitoring (SRM) mass spectrometry, to detect and assess RNAi-mediated depletion or "knockdown" of specific proteins from human cells and from Drosophila flies. This label-free approach does not require any specific reagents to confirm the depletion of RNAi target protein(s) in unfractionated cell or whole organism extracts. The protocol described here is general, can be developed rapidly, and can be multiplexed to detect and measure multiple proteins at once. Furthermore, the methodology can be extended to any tandem mass spectrometer, making it widely accessible. This methodology will be applicable to a wide range of basic science and clinical questions where RNAi-mediated protein depletion needs to be verified, or where differences in relative abundance of target proteins need to be rapidly assessed between samples.
Figures





References
-
- Mullenders J, Bernards R. Loss-of-function genetic screens as a tool to improve the diagnosis and treatment of cancer. Oncogene. 2009;28(50):4409–4420. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

