Phase-defined complete sequencing of the HLA genes by next-generation sequencing
- PMID: 23714642
- PMCID: PMC3671147
- DOI: 10.1186/1471-2164-14-355
Phase-defined complete sequencing of the HLA genes by next-generation sequencing
Abstract
Background: The human leukocyte antigen (HLA) region, the 3.8-Mb segment of the human genome at 6p21, has been associated with more than 100 different diseases, mostly autoimmune diseases. Due to the complex nature of HLA genes, there are difficulties in elucidating complete HLA gene sequences especially HLA gene haplotype structures by the conventional sequencing method. We propose a novel, accurate, and cost-effective method for generating phase-defined complete sequencing of HLA genes by using indexed multiplex next generation sequencing.
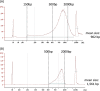
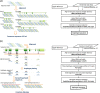
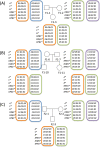
Results: A total of 33 HLA homozygous samples, 11 HLA heterozygous samples, and 3 parents-child families were subjected to phase-defined HLA gene sequencing. We applied long-range PCR to amplify six HLA genes (HLA-A, -C, -B, DRB1, -DQB1, and -DPB1) followed by transposase-based library construction and multiplex sequencing with the MiSeq sequencer. Paired-end reads (2 × 250 bp) derived from the sequencer were aligned to the six HLA gene segments of UCSC hg19 allowing at most 80 bases mismatch. For HLA homozygous samples, the six amplicons of an individual were pooled and simultaneously sequenced and mapped as an individual-tagging method. The paired-end reads were aligned to corresponding genes of UCSC hg19 and unambiguous, continuous sequences were obtained. For HLA heterozygous samples, each amplicon was separately sequenced and mapped as a gene-tagging method. After alignments, we detected informative paired-end reads harboring SNVs on both forward and reverse reads that are used to separate two chromosomes and to generate two phase-defined sequences in an individual. Consequently, we were able to determine the phase-defined HLA gene sequences from promoter to 3'-UTR and assign up to 8-digit HLA allele numbers, regardless of whether the alleles are rare or novel. Parent-child trio-based sequencing validated our sequencing and phasing methods.
Conclusions: Our protocol generated phased-defined sequences of the entire HLA genes, resulting in high resolution HLA typing and new allele detection.
Figures
References
-
- Shiina T, Hosomichi K, Inoko H, Kulski JK. The HLA genomic loci map: expression, interaction, diversity and disease. J Hum Genet. 2009;5:15–39. - PubMed
-
- Stewart CA, Horton R, Allcock RJ, Ashurst JL, Atrazhev AM, Coggill P, Dunham I, Forbes S, Halls K, Howson JM, Humphray SJ, Hunt S, Mungall AJ, Osoegawa K, Palmer S, Roberts AN, Rogers J, Sims S, Wang Y, Wilming LG, Elliott JF, de Jong PJ, Sawcer S, Todd JA, Trowsdale J, Beck S. Complete MHC haplotype sequencing for common disease gene mapping. Genome Res. 2004;14:1176–1187. doi: 10.1101/gr.2188104. - DOI - PMC - PubMed
-
- Horton R, Gibson R, Coggill P, Miretti M, Allcock RJ, Almeida J, Forbes S, Gilbert JG, Halls K, Harrow JL, Hart E, Howe K, Jackson DK, Palmer S, Roberts AN, Sims S, Stewart CA, Traherne JA, Trevanion S, Wilming L, Rogers J, de Jong PJ, Elliott JF, Sawcer S, Todd JA, Trowsdale J, Beck S. Variation analysis and gene annotation of eight MHC haplotypes: the MHC Haplotype Project. Immunogenetics. 2008;60:1–18. doi: 10.1007/s00251-007-0262-2. - DOI - PMC - PubMed
-
- Traherne JA, Horton R, Roberts AN, Miretti MM, Hurles ME, Stewart CA, Ashurst JL, Atrazhev AM, Coggill P, Palmer S, Almeida J, Sims S, Wilming LG, Rogers J, de Jong PJ, Carrington M, Elliott JF, Sawcer S, Todd JA, Trowsdale J, Beck S. Genetic analysis of completely sequenced disease-associated MHC haplotypes identifies shuffling of segments in recent human history. PLoS Genet. 2006;2:e9. doi: 10.1371/journal.pgen.0020009. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

