Metabolic and functional genomic studies identify deoxythymidylate kinase as a target in LKB1-mutant lung cancer
- PMID: 23715154
- PMCID: PMC3753578
- DOI: 10.1158/2159-8290.CD-13-0015
Metabolic and functional genomic studies identify deoxythymidylate kinase as a target in LKB1-mutant lung cancer
Abstract
The LKB1/STK11 tumor suppressor encodes a serine/threonine kinase, which coordinates cell growth, polarity, motility, and metabolism. In non-small cell lung carcinoma, LKB1 is somatically inactivated in 25% to 30% of cases, often concurrently with activating KRAS mutations. Here, we used an integrative approach to define novel therapeutic targets in KRAS-driven LKB1-mutant lung cancers. High-throughput RNA interference screens in lung cancer cell lines from genetically engineered mouse models driven by activated KRAS with or without coincident Lkb1 deletion led to the identification of Dtymk, encoding deoxythymidylate kinase (DTYMK), which catalyzes dTTP biosynthesis, as synthetically lethal with Lkb1 deficiency in mouse and human lung cancer lines. Global metabolite profiling showed that Lkb1-null cells had a striking decrease in multiple nucleotide metabolites as compared with the Lkb1-wild-type cells. Thus, LKB1-mutant lung cancers have deficits in nucleotide metabolism that confer hypersensitivity to DTYMK inhibition, suggesting that DTYMK is a potential therapeutic target in this aggressive subset of tumors.
Figures
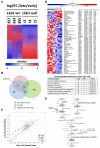
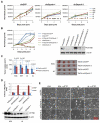
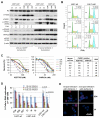

Comment in
-
Energizing the search to target LKB1-mutant tumors.Cancer Discov. 2013 Aug;3(8):843-5. doi: 10.1158/2159-8290.CD-13-0296. Cancer Discov. 2013. PMID: 23928772 Free PMC article.
References
-
- Wodarz A, Nathke I. Cell polarity in development and cancer. Nature cell biology. 2007;9:1016–24. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P50CA090578/CA/NCI NIH HHS/United States
- CA166480/CA/NCI NIH HHS/United States
- R01 CA122794/CA/NCI NIH HHS/United States
- R01 CA095175/CA/NCI NIH HHS/United States
- CA122794/CA/NCI NIH HHS/United States
- U01 CA141576/CA/NCI NIH HHS/United States
- P01 CA120964/CA/NCI NIH HHS/United States
- R01 GM073894/GM/NIGMS NIH HHS/United States
- CA163896/CA/NCI NIH HHS/United States
- P01 CA154303/CA/NCI NIH HHS/United States
- R01 CA142794/CA/NCI NIH HHS/United States
- R01 CA163896/CA/NCI NIH HHS/United States
- R01 CA166480/CA/NCI NIH HHS/United States
- R01 CA157490/CA/NCI NIH HHS/United States
- R01 CA140594/CA/NCI NIH HHS/United States
- CA140594/CA/NCI NIH HHS/United States
- R01 CA042802/CA/NCI NIH HHS/United States
- P50 CA090578/CA/NCI NIH HHS/United States
- CA154303/CA/NCI NIH HHS/United States
- K08 CA151651/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

