Drug-induced histone eviction from open chromatin contributes to the chemotherapeutic effects of doxorubicin
- PMID: 23715267
- PMCID: PMC3674280
- DOI: 10.1038/ncomms2921
Drug-induced histone eviction from open chromatin contributes to the chemotherapeutic effects of doxorubicin
Abstract
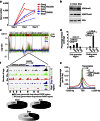
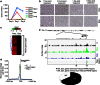
DNA topoisomerase II inhibitors are a major class of cancer chemotherapeutics, which are thought to eliminate cancer cells by inducing DNA double-strand breaks. Here we identify a novel activity for the anthracycline class of DNA topoisomerase II inhibitors: histone eviction from open chromosomal areas. We show that anthracyclines promote histone eviction irrespective of their ability to induce DNA double-strand breaks. The histone variant H2AX, which is a key component of the DNA damage response, is also evicted by anthracyclines, and H2AX eviction is associated with attenuated DNA repair. Histone eviction deregulates the transcriptome in cancer cells and organs such as the heart, and can drive apoptosis of topoisomerase-negative acute myeloid leukaemia blasts in patients. We define a novel mechanism of action of anthracycline anticancer drugs doxorubicin and daunorubicin on chromatin biology, with important consequences for DNA damage responses, epigenetics, transcription, side effects and cancer therapy.
Figures




References
-
- Farmer H. et al.. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434, 917–921 (2005). - PubMed
-
- Arcamone F. et al.. Adriamycin, 14-hydroxydaimomycin, a new antitumor antibiotic from S. Peucetius var. caesius. Biotechnol Bioeng. 11, 1101–1110 (1969). - PubMed
-
- Hande K. R. Clinical applications of anticancer drugs targeted to topoisomerase II. Biochim. Biophys. Acta. 1400, 173–184 (1998). - PubMed
-
- Minotti G., Menna P., Salvatorelli E., Cairo G. & Gianni L. Anthracyclines: molecular advances and pharmacologic developments in antitumor activity and cardiotoxicity. Pharmacol. Rev. 56, 185–229 (2004). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

