Compensation of the metabolic costs of antibiotic resistance by physiological adaptation in Escherichia coli
- PMID: 23716056
- PMCID: PMC3719774
- DOI: 10.1128/AAC.02096-12
Compensation of the metabolic costs of antibiotic resistance by physiological adaptation in Escherichia coli
Abstract
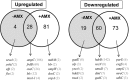
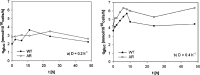
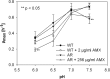
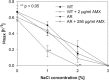
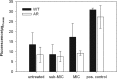
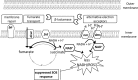
Antibiotic resistance is often associated with metabolic costs. To investigate the metabolic consequences of antibiotic resistance, the genomic and transcriptomic profiles of an amoxicillin-resistant Escherichia coli strain and the wild type it was derived from were compared. A total of 125 amino acid substitutions and 7 mutations that were located <1,000 bp upstream of differentially expressed genes were found in resistant cells. However, broad induction and suppression of genes were observed when comparing the expression profiles of resistant and wild-type cells. Expression of genes involved in cell wall maintenance, DNA metabolic processes, cellular stress response, and respiration was most affected in resistant cells regardless of the absence or presence of amoxicillin. The SOS response was downregulated in resistant cells. The physiological effect of the acquisition of amoxicillin resistance in cells grown in chemostat cultures consisted of an initial increase in glucose consumption that was followed by an adaptation process. Furthermore, no difference in maintenance energy was observed between resistant and sensitive cells. In accordance with the transcriptomic profile, exposure of resistant cells to amoxicillin resulted in reduced salt and pH tolerance. Taken together, the results demonstrate that the acquisition of antibiotic resistance in E. coli is accompanied by specifically reorganized metabolic networks in order to circumvent metabolic costs. The overall effect of the acquisition of resistance consists not so much of an extra energy requirement, but more a reduced ecological range.
Figures


References
-
- Andersson DI, Levin BR. 1999. The biological cost of antibiotic resistance. Curr. Opin. Microbiol. 2:489–493 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

