Human embryonic stem cell lines model experimental human cytomegalovirus latency
- PMID: 23716573
- PMCID: PMC3663570
- DOI: 10.1128/mBio.00298-13
Human embryonic stem cell lines model experimental human cytomegalovirus latency
Abstract
Herpesviruses are highly successful pathogens that persist for the lifetime of their hosts primarily because of their ability to establish and maintain latent infections from which the virus is capable of productively reactivating. Human cytomegalovirus (HCMV), a betaherpesvirus, establishes latency in CD34(+) hematopoietic progenitor cells during natural infections in the body. Experimental infection of CD34(+) cells ex vivo has demonstrated that expression of the viral gene products that drive productive infection is silenced by an intrinsic immune defense mediated by Daxx and histone deacetylases through heterochromatinization of the viral genome during the establishment of latency. Additional mechanistic details about the establishment, let alone maintenance and reactivation, of HCMV latency remain scarce. This is partly due to the technical challenges of CD34(+) cell culture, most notably, the difficulty in preventing spontaneous differentiation that drives reactivation and renders them permissive for productive infection. Here we demonstrate that HCMV can establish, maintain, and reactivate in vitro from experimental latency in cultures of human embryonic stem cells (ESCs), for which spurious differentiation can be prevented or controlled. Furthermore, we show that known molecular aspects of HCMV latency are faithfully recapitulated in these cells. In total, we present ESCs as a novel, tractable model for studies of HCMV latency.
Figures
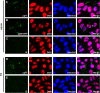
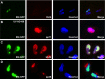
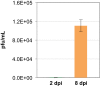

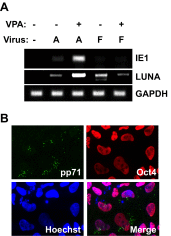
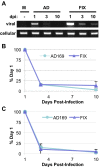

References
-
- Mocarski E, Shenk T, Pass R. 2007. Cytomegaloviruses, p 2701–2772 In Knipe D, Howley P, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE, Fields virology. Lippincott Williams and Wilkins, Philadelphia, PA.
-
- Marschall M, Stamminger T. 2009. Molecular targets for antiviral therapy of cytomegalovirus infections. Future Microbiol. 4:731–742 - PubMed
-
- Reeves M, Sinclair J. 2008. Aspects of human cytomegalovirus latency and reactivation. Curr. Top. Microbiol. Immunol. 325:297–313 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
